Multi-tissue transcriptomic study reveals the main role of liver in the chicken adaptive response to a switch in dietary energy source through the transcriptional regulation of lipogenesis
- PMID: 29514634
- PMCID: PMC5842524
- DOI: 10.1186/s12864-018-4520-5
Multi-tissue transcriptomic study reveals the main role of liver in the chicken adaptive response to a switch in dietary energy source through the transcriptional regulation of lipogenesis
Abstract
Background: Because the cost of cereals is unstable and represents a large part of production charges for meat-type chicken, there is an urge to formulate alternative diets from more cost-effective feedstuff. We have recently shown that meat-type chicken source is prone to adapt to dietary starch substitution with fat and fiber. The aim of this study was to better understand the molecular mechanisms of this adaptation to changes in dietary energy sources through the fine characterization of transcriptomic changes occurring in three major metabolic tissues - liver, adipose tissue and muscle - as well as in circulating blood cells.
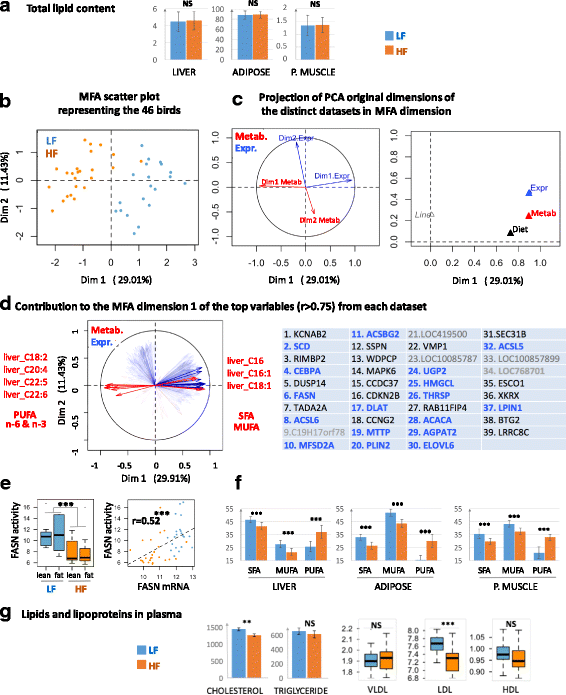
Results: We revealed the fine-tuned regulation of many hepatic genes encoding key enzymes driving glycogenesis and de novo fatty acid synthesis pathways and of some genes participating in oxidation. Among the genes expressed upon consumption of a high-fat, high-fiber diet, we highlighted CPT1A, which encodes a key enzyme in the regulation of fatty acid oxidation. Conversely, the repression of lipogenic genes by the high-fat diet was clearly associated with the down-regulation of SREBF1 transcripts but was not associated with the transcript regulation of MLXIPL and NR1H3, which are both transcription factors. This result suggests a pivotal role for SREBF1 in lipogenesis regulation in response to a decrease in dietary starch and an increase in dietary PUFA. Other prospective regulators of de novo hepatic lipogenesis were suggested, such as PPARD, JUN, TADA2A and KAT2B, the last two genes belonging to the lysine acetyl transferase (KAT) complex family regulating histone and non-histone protein acetylation. Hepatic glycogenic genes were also down-regulated in chickens fed a high-fat, high-fiber diet compared to those in chickens fed a starch-based diet. No significant dietary-associated variations in gene expression profiles was observed in the other studied tissues, suggesting that the liver mainly contributed to the adaptation of birds to changes in energy source and nutrients in their diets, at least at the transcriptional level. Moreover, we showed that PUFA deposition observed in the different tissues may not rely on transcriptional changes.
Conclusion: We showed the major role of the liver, at the gene expression level, in the adaptive response of chicken to dietary starch substitution with fat and fiber.
Keywords: Adaptation; Chicken; Fat diet; Gene expression; Lipid; PUFA; Regulation; SREBF1; TADA2A.
Conflict of interest statement
Ethics approval and consent to participate
All experiments were conducted following the French National Guidelines for the care and use of animals in research edited by the French Ministries of Higher Education and Scientific Research, and of Agriculture and Fisheries
Consent for publication
“Not applicable”
Competing interests
The authors declare that they have competing interests with Elisabeth Lebihan as a member of the editorial board of BMC genomics journal.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures






References
-
- Baéza E, Gondret F, Chartrin P, Le Bihan-Duval E, Berri C, Gabriel I, et al. The ability of genetically lean or fat slow-growing chickens to synthesize and store lipids is not altered by the dietary energy source. Anim Int J Anim Biosci. 2015;9:1643–1652. doi: 10.1017/S1751731115000683. - DOI - PubMed
-
- Gondret F, Louveau I, Mourot J, Duclos MJ, Lagarrigue S, Gilbert H, et al. Dietary energy sources affect the partition of body lipids and the hierarchy of energy metabolic pathways in growing pigs differing in feed efficiency. J Anim Sci. 2014;92:4865–4877. doi: 10.2527/jas.2014-7995. - DOI - PubMed
-
- Désert C, Merlot E, Zerjal T, Bed’hom B, Härtle S, Le Cam A, et al. Transcriptomes of whole blood and PBMC in chickens. Comp Biochem Physiol. 2016;20:1–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

