Phosphatidylinositol 3-Kinase-DNA Methyltransferase 1-miR-1281-Histone Deacetylase 4 Regulatory Axis Mediates Platelet-Derived Growth Factor-Induced Proliferation and Migration of Pulmonary Artery Smooth Muscle Cells
- PMID: 29514810
- PMCID: PMC5907547
- DOI: 10.1161/JAHA.117.007572
Phosphatidylinositol 3-Kinase-DNA Methyltransferase 1-miR-1281-Histone Deacetylase 4 Regulatory Axis Mediates Platelet-Derived Growth Factor-Induced Proliferation and Migration of Pulmonary Artery Smooth Muscle Cells
Abstract
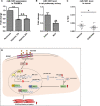
Background: Platelet-derived growth factor BB, a potent mitogen of pulmonary artery smooth muscle cells (PASMCs), has been implicated in pulmonary arterial remodeling, which is a key pathogenic feature of pulmonary arterial hypertension. Previous microRNA profiling in platelet-derived growth factor BB-treated PASMCs found a significantly downregulated microRNA, miR-1281, but it has not been associated with any cellular function, and we investigated the possibility.
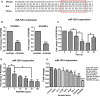
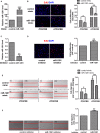
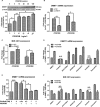
Methods and results: Real-time quantitative reverse transcription-polymerase chain reaction assay proved that downregulation of miR-1281 was a conserved phenomenon in human and rat PASMCs. Overexpression and inhibition of miR-1281 in PASMCs promoted and suppressed, respectively, the cell proliferation and migration. Bioinformatic prediction and 3'-untranslated region reporter assay identified histone deacetylase 4 to be a direct target of miR-1281. Supporting this, proliferation and migration assay demonstrated the cellular function of histone deacetylase 4 is inversely correlated with that of miR-1281. Mechanistically, it is found that platelet-derived growth factor BB activates the phosphatidylinositol 3-kinase pathway, which then induces the expression of DNA methyltransferase 1, leading to enhanced methylation of a flanking CpG island and repressed miR-1281 expression. Finally, a reduced miR-1281 level was consistently identified in hypoxic PASMCs in vitro, in pulmonary arteries of rats with monocrotaline-induced pulmonary arterial hypertension, and in serum of patients with coronary heart disease-pulmonary arterial hypertension. These data suggest that there may be a diagnostic and therapeutic use for miR-1281.
Conclusions: Herein, we report a novel regulatory axis, phosphatidylinositol 3-kinase-DNA methyltransferase 1-miR-1281-histone deacetylase 4, integrating multiple epigenetic regulators that participate in platelet-derived growth factor BB-stimulated PASMC proliferation and migration and pulmonary vascular remodeling.
Keywords: DNA methyltransferase 1; epigenetics; histone deacetylase 4; hypertension; miRNA; platelet‐derived growth factor; pulmonary; pulmonary arterial smooth muscle cells; vascular remodeling; vascular smooth muscle.
© 2018 The Authors. Published on behalf of the American Heart Association, Inc., by Wiley.
Figures

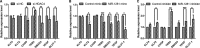
Similar articles
-
PDGFBB promotes proliferation and migration via regulating miR-1181/STAT3 axis in human pulmonary arterial smooth muscle cells.Am J Physiol Lung Cell Mol Physiol. 2018 Dec 1;315(6):L965-L976. doi: 10.1152/ajplung.00224.2018. Epub 2018 Sep 13. Am J Physiol Lung Cell Mol Physiol. 2018. PMID: 30211651
-
LncRNA-SMILR modulates RhoA/ROCK signaling by targeting miR-141 to regulate vascular remodeling in pulmonary arterial hypertension.Am J Physiol Heart Circ Physiol. 2020 Aug 1;319(2):H377-H391. doi: 10.1152/ajpheart.00717.2019. Epub 2020 Jun 19. Am J Physiol Heart Circ Physiol. 2020. PMID: 32559140
-
lncRNA VELRP Modulates Pulmonary Arterial Smooth Muscle Cell Proliferation and Promotes Vascular Remodeling in Pulmonary Hypertension.Arterioscler Thromb Vasc Biol. 2024 Dec;44(12):2560-2576. doi: 10.1161/ATVBAHA.124.321416. Epub 2024 Oct 3. Arterioscler Thromb Vasc Biol. 2024. PMID: 39360410
-
MicroRNAs in pulmonary arterial remodeling.Cell Mol Life Sci. 2013 Dec;70(23):4479-94. doi: 10.1007/s00018-013-1382-5. Epub 2013 Jun 6. Cell Mol Life Sci. 2013. PMID: 23739951 Free PMC article. Review.
-
MicroRNA in pulmonary vascular disease.Prog Mol Biol Transl Sci. 2014;124:43-63. doi: 10.1016/B978-0-12-386930-2.00003-3. Prog Mol Biol Transl Sci. 2014. PMID: 24751426 Review.
Cited by
-
Non-Coding RNA Networks in Pulmonary Hypertension.Front Genet. 2021 Nov 30;12:703860. doi: 10.3389/fgene.2021.703860. eCollection 2021. Front Genet. 2021. PMID: 34917122 Free PMC article. Review.
-
The Role of Clonal Hematopoiesis of Indeterminant Potential and DNA (Cytosine-5)-Methyltransferase Dysregulation in Pulmonary Arterial Hypertension and Other Cardiovascular Diseases.Cells. 2023 Oct 26;12(21):2528. doi: 10.3390/cells12212528. Cells. 2023. PMID: 37947606 Free PMC article. Review.
-
MiR-1281 is involved in depression disorder and the antidepressant effects of Kai-Xin-San by targeting ADCY1 and DVL1.Heliyon. 2023 Mar 7;9(3):e14265. doi: 10.1016/j.heliyon.2023.e14265. eCollection 2023 Mar. Heliyon. 2023. PMID: 36938448 Free PMC article.
-
MicroRNA Nanotherapeutics for Lung Targeting. Insights into Pulmonary Hypertension.Int J Mol Sci. 2020 May 4;21(9):3253. doi: 10.3390/ijms21093253. Int J Mol Sci. 2020. PMID: 32375361 Free PMC article. Review.
-
Epigenetic Mechanisms as Emerging Therapeutic Targets and Microfluidic Chips Application in Pulmonary Arterial Hypertension.Biomedicines. 2022 Jan 13;10(1):170. doi: 10.3390/biomedicines10010170. Biomedicines. 2022. PMID: 35052850 Free PMC article. Review.
References
-
- Ten Freyhaus H, Berghausen EM, Janssen W, Leuchs M, Zierden M, Murmann K, Klinke A, Vantler M, Caglayan E, Kramer T, Baldus S, Schermuly RT, Tallquist MD, Rosenkranz S. Genetic ablation of PDGF‐dependent signaling pathways abolishes vascular remodeling and experimental pulmonary hypertension. Arterioscler Thromb Vasc Biol. 2015;35:1236–1245. - PMC - PubMed
-
- Hoeper MM, Huscher D, Ghofrani HA, Delcroix M, Distler O, Schweiger C, Grunig E, Staehler G, Rosenkranz S, Halank M. Elderly patients diagnosed with idiopathic pulmonary arterial hypertension: results from the COMPERA registry. Int J Cardiol. 2013;168:871–880. - PubMed
-
- Humbert M, Sitbon O, Chaouat A, Bertocchi M, Habib G, Gressin V, Yaïci A, Weitzenblum E, Cordier J‐F, Chabot F. Survival in patients with idiopathic, familial, and anorexigen‐associated pulmonary arterial hypertension in the modern management era. Circulation. 2010;122:156–163. - PubMed
-
- Benza RL, Miller DP, Gomberg‐Maitland M, Frantz RP, Foreman AJ, Coffey CS, Frost A, Barst RJ, Badesch DB, Elliott CG. Predicting survival in pulmonary arterial hypertension insights from the registry to evaluate early and long‐term pulmonary arterial hypertension disease management (REVEAL). Circulation. 2010;122:164–172. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

