A Stem-Loop Structure in Potato Leafroll Virus Open Reading Frame 5 (ORF5) Is Essential for Readthrough Translation of the Coat Protein ORF Stop Codon 700 Bases Upstream
- PMID: 29514911
- PMCID: PMC5952135
- DOI: 10.1128/JVI.01544-17
A Stem-Loop Structure in Potato Leafroll Virus Open Reading Frame 5 (ORF5) Is Essential for Readthrough Translation of the Coat Protein ORF Stop Codon 700 Bases Upstream
Abstract
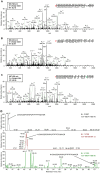
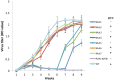
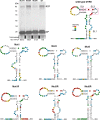
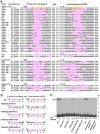
Translational readthrough of the stop codon of the capsid protein (CP) open reading frame (ORF) is used by members of the Luteoviridae to produce their minor capsid protein as a readthrough protein (RTP). The elements regulating RTP expression are not well understood, but they involve long-distance interactions between RNA domains. Using high-resolution mass spectrometry, glutamine and tyrosine were identified as the primary amino acids inserted at the stop codon of Potato leafroll virus (PLRV) CP ORF. We characterized the contributions of a cytidine-rich domain immediately downstream and a branched stem-loop structure 600 to 700 nucleotides downstream of the CP stop codon. Mutations predicted to disrupt and restore the base of the distal stem-loop structure prevented and restored stop codon readthrough. Motifs in the downstream readthrough element (DRTE) are predicted to base pair to a site within 27 nucleotides (nt) of the CP ORF stop codon. Consistent with a requirement for this base pairing, the DRTE of Cereal yellow dwarf virus was not compatible with the stop codon-proximal element of PLRV in facilitating readthrough. Moreover, deletion of the complementary tract of bases from the stop codon-proximal region or the DRTE of PLRV prevented readthrough. In contrast, the distance and sequence composition between the two domains was flexible. Mutants deficient in RTP translation moved long distances in plants, but fewer infection foci developed in systemically infected leaves. Selective 2'-hydroxyl acylation and primer extension (SHAPE) probing to determine the secondary structure of the mutant DRTEs revealed that the functional mutants were more likely to have bases accessible for long-distance base pairing than the nonfunctional mutants. This study reveals a heretofore unknown combination of RNA structure and sequence that reduces stop codon efficiency, allowing translation of a key viral protein.IMPORTANCE Programmed stop codon readthrough is used by many animal and plant viruses to produce key viral proteins. Moreover, such "leaky" stop codons are used in host mRNAs or can arise from mutations that cause genetic disease. Thus, it is important to understand the mechanism(s) of stop codon readthrough. Here, we shed light on the mechanism of readthrough of the stop codon of the coat protein ORFs of viruses in the Luteoviridae by identifying the amino acids inserted at the stop codon and RNA structures that facilitate this "leakiness" of the stop codon. Members of the Luteoviridae encode a C-terminal extension to the capsid protein known as the readthrough protein (RTP). We characterized two RNA domains in Potato leafroll virus (PLRV), located 600 to 700 nucleotides apart, that are essential for efficient RTP translation. We further determined that the PLRV readthrough process involves both local structures and long-range RNA-RNA interactions. Genetic manipulation of the RNA structure altered the ability of PLRV to translate RTP and systemically infect the plant. This demonstrates that plant virus RNA contains multiple layers of information beyond the primary sequence and extends our understanding of stop codon readthrough. Strategic targets that can be exploited to disrupt the virus life cycle and reduce its ability to move within and between plant hosts were revealed.
Keywords: RNA structure; polerovirus; readthrough; systemic infection; translational control.
Copyright © 2018 American Society for Microbiology.
Figures



Similar articles
-
An aromatic amino acid and associated helix in the C-terminus of the potato leafroll virus minor capsid protein regulate systemic infection and symptom expression.PLoS Pathog. 2018 Nov 15;14(11):e1007451. doi: 10.1371/journal.ppat.1007451. eCollection 2018 Nov. PLoS Pathog. 2018. PMID: 30440046 Free PMC article.
-
Complex and simple translational readthrough signals in pea enation mosaic virus 1 and potato leafroll virus, respectively.PLoS Pathog. 2022 Sep 29;18(9):e1010888. doi: 10.1371/journal.ppat.1010888. eCollection 2022 Sep. PLoS Pathog. 2022. PMID: 36174104 Free PMC article.
-
Small deletions in the potato leafroll virus readthrough protein affect particle morphology, aphid transmission, virus movement and accumulation.J Gen Virol. 2008 Aug;89(Pt 8):2037-2045. doi: 10.1099/vir.0.83625-0. J Gen Virol. 2008. PMID: 18632976
-
Translational termination-reinitiation in RNA viruses.Biochem Soc Trans. 2010 Dec;38(6):1558-64. doi: 10.1042/BST0381558. Biochem Soc Trans. 2010. PMID: 21118126 Review.
-
Functional analysis of structural motifs in dicistroviruses.Virus Res. 2009 Feb;139(2):137-47. doi: 10.1016/j.virusres.2008.06.006. Epub 2008 Jul 25. Virus Res. 2009. PMID: 18621089 Review.
Cited by
-
Genetic Diversity Analysis of Brassica Yellows Virus Causing Aberrant Color Symptoms in Oilseed Rape.Plants (Basel). 2023 Feb 23;12(5):1008. doi: 10.3390/plants12051008. Plants (Basel). 2023. PMID: 36903869 Free PMC article.
-
Bioluminescence Production by Turnip Yellows Virus Infectious Clones: A New Way to Monitor Plant Virus Infection.Int J Mol Sci. 2022 Nov 8;23(22):13685. doi: 10.3390/ijms232213685. Int J Mol Sci. 2022. PMID: 36430165 Free PMC article.
-
RNA-mediated translation regulation in viral genomes: computational advances in the recognition of sequences and structures.Brief Bioinform. 2020 Jul 15;21(4):1151-1163. doi: 10.1093/bib/bbz054. Brief Bioinform. 2020. PMID: 31204430 Free PMC article. Review.
-
Polerovirus genomic variation.Virus Evol. 2021 Dec 4;7(2):veab102. doi: 10.1093/ve/veab102. eCollection 2021 Sep. Virus Evol. 2021. PMID: 35299789 Free PMC article.
-
The Interaction Dynamics of Two Potato Leafroll Virus Movement Proteins Affects Their Localization to the Outer Membranes of Mitochondria and Plastids.Viruses. 2018 Oct 26;10(11):585. doi: 10.3390/v10110585. Viruses. 2018. PMID: 30373157 Free PMC article.
References
-
- Newburn LR, White KA. 2015. Cis-acting RNA elements in positive-strand RNA plant virus genomes. Virology 479–480:434–443. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

