CLAMP/Spef1 regulates planar cell polarity signaling and asymmetric microtubule accumulation in the Xenopus ciliated epithelia
- PMID: 29514918
- PMCID: PMC5940297
- DOI: 10.1083/jcb.201706058
CLAMP/Spef1 regulates planar cell polarity signaling and asymmetric microtubule accumulation in the Xenopus ciliated epithelia
Abstract
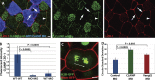
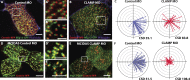
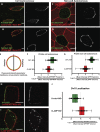
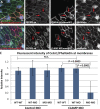
Most epithelial cells polarize along the axis of the tissue, a feature known as planar cell polarity (PCP). The initiation of PCP requires cell-cell signaling via the noncanonical Wnt/PCP pathway. Additionally, changes in the cytoskeleton both facilitate and reflect this polarity. We have identified CLAMP/Spef1 as a novel regulator of PCP signaling. In addition to decorating microtubules (MTs) and the ciliary rootlet, a pool of CLAMP localizes at the apical cell cortex. Depletion of CLAMP leads to the loss of PCP protein asymmetry, defects in cilia polarity, and defects in the angle of cell division. Additionally, depletion of CLAMP leads to a loss of the atypical cadherin-like molecule Celrs2, suggesting that CLAMP facilitates the stabilization of junctional interactions responsible for proper PCP protein localization. Depletion of CLAMP also affects the polarized organization of MTs. We hypothesize that CLAMP facilitates the establishment of cell polarity and promotes the asymmetric accumulation of MTs downstream of the establishment of proper PCP.
© 2018 Kim et al.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

