Cell size-dependent regulation of Wee1 localization by Cdr2 cortical nodes
- PMID: 29514920
- PMCID: PMC5940308
- DOI: 10.1083/jcb.201709171
Cell size-dependent regulation of Wee1 localization by Cdr2 cortical nodes
Abstract
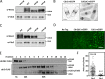
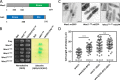
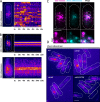
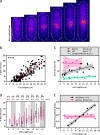
Cell size control requires mechanisms that link cell growth with Cdk1 activity. In fission yeast, the protein kinase Cdr2 forms cortical nodes that include the Cdk1 inhibitor Wee1 along with the Wee1-inhibitory kinase Cdr1. We investigated how nodes inhibit Wee1 during cell growth. Biochemical fractionation revealed that Cdr2 nodes were megadalton structures enriched for activated Cdr2, which increases in level during interphase growth. In live-cell total internal reflection fluorescence microscopy videos, Cdr2 and Cdr1 remained constant at nodes over time, but Wee1 localized to nodes in short bursts. Recruitment of Wee1 to nodes required Cdr2 kinase activity and the noncatalytic N terminus of Wee1. Bursts of Wee1 localization to nodes increased 20-fold as cells doubled in size throughout G2. Size-dependent signaling was caused in part by the Cdr2 inhibitor Pom1, which suppressed Wee1 node bursts in small cells. Thus, increasing Cdr2 activity during cell growth promotes Wee1 localization to nodes, where inhibitory phosphorylation of Wee1 by Cdr1 and Cdr2 kinases promotes mitotic entry.
© 2018 Allard et al.
Figures
References
-
- Bähler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A. III, Steever A.B., Wach A., Philippsen P., and Pringle J.R.. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14:943–951. 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y - DOI - PubMed
-
- Bhatia P., Hachet O., Hersch M., Rincon S.A., Berthelot-Grosjean M., Dalessi S., Basterra L., Bergmann S., Paoletti A., and Martin S.G.. 2014. Distinct levels in Pom1 gradients limit Cdr2 activity and localization to time and position division. Cell Cycle. 13:538–552. 10.4161/cc.27411 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

