RecQ Helicases Function in Development, DNA Repair, and Gene Targeting in Physcomitrella patens
- PMID: 29514942
- PMCID: PMC5894843
- DOI: 10.1105/tpc.17.00632
RecQ Helicases Function in Development, DNA Repair, and Gene Targeting in Physcomitrella patens
Abstract
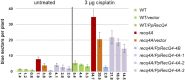
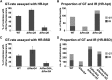
RecQ DNA helicases are genome surveillance proteins found in all kingdoms of life. They are characterized best in humans, as mutations in RecQ genes lead to developmental abnormalities and diseases. To better understand RecQ functions in plants we concentrated on Arabidopsis thaliana and Physcomitrella patens, the model species predominantly used for studies on DNA repair and gene targeting. Phylogenetic analysis of the six P. patens RecQ genes revealed their orthologs in humans and plants. Because Arabidopsis and P. patens differ in their RecQ4 and RecQ6 genes, reporter and deletion moss mutants were generated and gene functions studied in reciprocal cross-species and cross-kingdom approaches. Both proteins can be found in meristematic moss tissues, although at low levels and with distinct expression patterns. PpRecQ4 is involved in embryogenesis and in subsequent development as demonstrated by sterility of ΔPpRecQ4 mutants and by morphological aberrations. Additionally, ΔPpRecQ4 displays an increased sensitivity to DNA damages and an increased rate of gene targeting. Therefore, we conclude that PpRecQ4 acts as a repressor of recombination. In contrast, PpRecQ6 is not obviously important for moss development or DNA repair but does function as a potent enhancer of gene targeting.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures







References
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410. - PubMed
-
- Bagherieh-Najjar M.B., de Vries O.M.H., Kroon J.T.M., Wright E.L., Elborough K.M., Hille J., Dijkwel P.P. (2003). Arabidopsis RecQsim, a plant-specific member of the RecQ helicase family, can suppress the MMS hypersensitivity of the yeast sgs1 mutant. Plant Mol. Biol. 52: 273–284. - PubMed
-
- Bagherieh-Najjar M.B., de Vries O.M.H., Hille J., Dijkwel P.P. (2005). Arabidopsis RecQI4A suppresses homologous recombination and modulates DNA damage responses. Plant J. 43: 789–798. - PubMed
-
- Baumann P., West S.C. (1998). Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem. Sci. 23: 247–251. - PubMed
-
- Beike A.K., von Stackelberg M., Schallenberg-Rüdinger M., Hanke S.T., Follo M., Quandt D., McDaniel S.F., Reski R., Tan B.C., Rensing S.A. (2014). Molecular evidence for convergent evolution and allopolyploid speciation within the Physcomitrium-Physcomitrella species complex. BMC Evol. Biol. 14: 158. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

