Intra-species Genomic and Physiological Variability Impact Stress Resistance in Strains of Probiotic Potential
- PMID: 29515537
- PMCID: PMC5826259
- DOI: 10.3389/fmicb.2018.00242
Intra-species Genomic and Physiological Variability Impact Stress Resistance in Strains of Probiotic Potential
Abstract
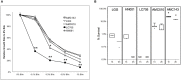
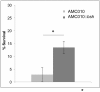
Large-scale microbiome studies have established that most of the diversity contained in the gastrointestinal tract is represented at the strain level; however, exhaustive genomic and physiological characterization of human isolates is still lacking. With increased use of probiotics as interventions for gastrointestinal disorders, genomic and functional characterization of novel microorganisms becomes essential. In this study, we explored the impact of strain-level genomic variability on bacterial physiology of two novel human Lactobacillus rhamnosus strains (AMC143 and AMC010) of probiotic potential in relation to stress resistance. The strains showed differences with known probiotic strains (L. rhamnosus GG, Lc705, and HN001) at the genomic level, including nucleotide polymorphisms, mutations in non-coding regulatory regions, and rearrangements of genomic architecture. Transcriptomics analysis revealed that gene expression profiles differed between strains when exposed to simulated gastrointestinal stresses, suggesting the presence of unique regulatory systems in each strain. In vitro physiological assays to test resistance to conditions mimicking the gut environment (acid, alkali, and bile stress) showed that growth of L. rhamnosus AMC143 was inhibited upon exposure to alkaline pH, while AMC010 and control strain LGG were unaffected. AMC143 also showed a significant survival advantage compared to the other strains upon bile exposure. Reverse transcription qPCR targeting the bile salt hydrolase gene (bsh) revealed that AMC143 expressed bsh poorly (a consequence of a deletion in the bsh promoter and truncation of bsh gene in AMC143), while AMC010 had significantly higher expression levels than AMC143 or LGG. Insertional inactivation of the bsh gene in AMC010 suggested that bsh could be detrimental to bacterial survival during bile stress. Together, these findings show that coupling of classical microbiology with functional genomics methods for the characterization of bacterial strains is critical for the development of novel probiotics, as variability between strains can dramatically alter bacterial physiology and functionality.
Keywords: Lactobacillus rhamnosus; bacterial stress; bile salt hydrolase (BSH); intra-Species variability; probiotics.
Figures




References
-
- Anderson K. L., Roux C. M., Olson M. W., Luong T. T., Lee C. Y., Olson R., et al. (2010). Characterizing the effects of inorganic acid and Alkaline Shock on the Staphylococcus aureus transcriptome and messenger RNA turnover. FEMS Immunol. Med. Microbiol. 60, 208–250. 10.1111/j.1574-695X.2010.00736.x - DOI - PMC - PubMed
-
- Andreichin M. A. (1980). [Antimicrobial properties of bile and bile acids]. Antibiotiki 25, 936–939. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

