Evolution of Phototrophy in the Chloroflexi Phylum Driven by Horizontal Gene Transfer
- PMID: 29515543
- PMCID: PMC5826079
- DOI: 10.3389/fmicb.2018.00260
Evolution of Phototrophy in the Chloroflexi Phylum Driven by Horizontal Gene Transfer
Abstract
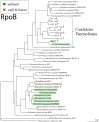
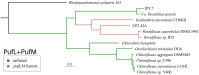
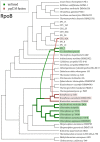
The evolutionary mechanisms behind the extant distribution of photosynthesis is a point of substantial contention. Hypotheses range from the presence of phototrophy in the last universal common ancestor and massive gene loss in most lineages, to a later origin in Cyanobacteria followed by extensive horizontal gene transfer into the extant phototrophic clades, with intermediate scenarios that incorporate aspects of both end-members. Here, we report draft genomes of 11 Chloroflexi: the phototrophic Chloroflexia isolate Kouleothrix aurantiaca as well as 10 genome bins recovered from metagenomic sequencing of microbial mats found in Japanese hot springs. Two of these metagenome bins encode photrophic reaction centers and several of these bins form a metabolically diverse, monophyletic clade sister to the Anaerolineae class that we term Candidatus Thermofonsia. Comparisons of organismal (based on conserved ribosomal) and phototrophy (reaction center and bacteriochlorophyll synthesis) protein phylogenies throughout the Chloroflexi demonstrate that two new lineages acquired phototrophy independently via horizontal gene transfer (HGT) from different ancestral donors within the classically phototrophic Chloroflexia class. These results illustrate a complex history of phototrophy within this group, with metabolic innovation tied to HGT. These observations do not support simple hypotheses for the evolution of photosynthesis that require massive character loss from many clades; rather, HGT appears to be the defining mechanic for the distribution of phototrophy in many of the extant clades in which it appears.
Keywords: comparative genomics; lateral gene transfer; microbial diversity; microbial metabolism; phylogenetics.
Figures
Similar articles
-
Microbial mats in the Turks and Caicos Islands reveal diversity and evolution of phototrophy in the Chloroflexota order Aggregatilineales.Environ Microbiome. 2020 Apr 16;15(1):9. doi: 10.1186/s40793-020-00357-8. Environ Microbiome. 2020. PMID: 33902735 Free PMC article.
-
Characterization of the First Cultured Representative of "Candidatus Thermofonsia" Clade 2 within Chloroflexi Reveals Its Phototrophic Lifestyle.mBio. 2022 Apr 26;13(2):e0028722. doi: 10.1128/mbio.00287-22. Epub 2022 Mar 1. mBio. 2022. PMID: 35229635 Free PMC article.
-
Evolutionary Implications of Anoxygenic Phototrophy in the Bacterial Phylum Candidatus Eremiobacterota (WPS-2).Front Microbiol. 2019 Jul 23;10:1658. doi: 10.3389/fmicb.2019.01658. eCollection 2019. Front Microbiol. 2019. PMID: 31396180 Free PMC article.
-
Prokaryotic photosynthesis and phototrophy illuminated.Trends Microbiol. 2006 Nov;14(11):488-96. doi: 10.1016/j.tim.2006.09.001. Epub 2006 Sep 25. Trends Microbiol. 2006. PMID: 16997562 Review.
-
Powered by light: Phototrophy and photosynthesis in prokaryotes and its evolution.Microbiol Res. 2016 May-Jun;186-187:99-118. doi: 10.1016/j.micres.2016.04.001. Epub 2016 Apr 2. Microbiol Res. 2016. PMID: 27242148 Review.
Cited by
-
Microbial mats in the Turks and Caicos Islands reveal diversity and evolution of phototrophy in the Chloroflexota order Aggregatilineales.Environ Microbiome. 2020 Apr 16;15(1):9. doi: 10.1186/s40793-020-00357-8. Environ Microbiome. 2020. PMID: 33902735 Free PMC article.
-
The Landscape of Genetic Content in the Gut and Oral Human Microbiome.Cell Host Microbe. 2019 Aug 14;26(2):283-295.e8. doi: 10.1016/j.chom.2019.07.008. Cell Host Microbe. 2019. PMID: 31415755 Free PMC article. Review.
-
Draft Genome Sequences of Two Basal Members of the Anaerolineae Class of Chloroflexi from a Sulfidic Hot Spring.Genome Announc. 2018 Jun 21;6(25):e00570-18. doi: 10.1128/genomeA.00570-18. Genome Announc. 2018. PMID: 29930070 Free PMC article.
-
Complex History of Aerobic Respiration and Phototrophy in the Chloroflexota Class Anaerolineae Revealed by High-Quality Draft Genome of Ca. Roseilinea mizusawaensis AA3_104.Microbes Environ. 2021;36(3):ME21020. doi: 10.1264/jsme2.ME21020. Microbes Environ. 2021. PMID: 34470945 Free PMC article.
-
What We Are Learning from the Diverse Structures of the Homodimeric Type I Reaction Center-Photosystems of Anoxygenic Phototropic Bacteria.Biomolecules. 2024 Mar 6;14(3):311. doi: 10.3390/biom14030311. Biomolecules. 2024. PMID: 38540731 Free PMC article. Review.
References
-
- Beer M., Seviour E. M., Kong Y., Cunningham M., Blackall L. L., Seviour R. J. (2002). Phylogeny of the filamentous bacterium Eikelboom Type 1851, and design and application of a 16S rRNA targeted oligonucleotide probe for its fluorescence in situ identification in activated sludge. FEMS Microbiol. Lett. 207, 179–183. 10.1111/j.1574-6968.2002.tb11048.x - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

