Functional Analysis of a Putative Type III Secretion System in Stress Adaption by Mesorhizobium alhagi CCNWXJ12-2T
- PMID: 29515545
- PMCID: PMC5826200
- DOI: 10.3389/fmicb.2018.00263
Functional Analysis of a Putative Type III Secretion System in Stress Adaption by Mesorhizobium alhagi CCNWXJ12-2T
Abstract
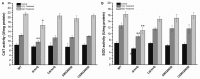
Mesorhizobium alhagi CCNWXJ12-2T, isolated from root nodules of the desert plant Alhagi sparsifolia, contains two type III secretion systems (T3SSs). T3SSs are specialized machinery with wide distribution in bacteria that inject effector proteins into target cells. Our previous study showed that the expression of M. alhagi T3SS1 is upregulated in high-salt conditions. Here, phylogenetic analysis of T3SS1 using the core protein RhcU suggested that T3SS1 belongs to the α-Rhc II subgroup of the Rhc T3SS family. To elaborate the function of M. alhagi CCNWXJ12-2T T3SS1 in stress adaption, two T3SS1 mutants (ΔrhcQ and ΔMA29250) were constructed and analyzed. β-galactosidase transcriptional fusion assays showed that activity of the promoter of T3SS1 was induced by salts. Mutant ΔrhcQ was more sensitive to NaCl and LiCl than the wild-type, but ΔMA29250 was not. Both mutants were more sensitive to KCl than the wild-type. The intracellular Na+ concentration in ΔrhcQ in high-NaCl conditions (0.4 M) increased by 37% compared to that of the wild-type strain, while the Na+ concentration in ΔMA29250 increased by 13%. The K+ concentration in both mutants increased by 16% compared to the wild-type in high-KCl conditions (0.3 M). Strain ΔrhcQ showed decreased survival compared to the wild-type after treatment with H2O2, while the survival rate of ΔMA29250 was almost the same as that of the wild-type. Antioxidant enzyme activities in ΔrhcQ were lower than those in the wild-type strain, but this was not the case for ΔMA29250. Our data elucidate the beneficial effects of T3SS1 in the adaption of M. alhagi CCNWXJ12-2T to stress.
Keywords: Mesorhizobium alhagi; Na+ content measurement; stress adaption; type III secretion system; β-galactosidase assay.
Figures






Similar articles
-
Role of exopolysaccharide in salt stress resistance and cell motility of Mesorhizobium alhagi CCNWXJ12-2T.Appl Microbiol Biotechnol. 2017 Apr;101(7):2967-2978. doi: 10.1007/s00253-017-8114-y. Epub 2017 Jan 17. Appl Microbiol Biotechnol. 2017. PMID: 28097405
-
Functional analysis of PrkA - a putative serine protein kinase from Mesorhizobium alhagi CCNWXJ12-2 - in stress resistance.BMC Microbiol. 2016 Sep 29;16(1):227. doi: 10.1186/s12866-016-0849-6. BMC Microbiol. 2016. PMID: 27686068 Free PMC article.
-
Global transcriptome analysis of Mesorhizobium alhagi CCNWXJ12-2 under salt stress.BMC Microbiol. 2014 Dec 24;14:1. doi: 10.1186/s12866-014-0319-y. BMC Microbiol. 2014. PMID: 25539655 Free PMC article.
-
Alhagi: a plant genus rich in bioactives for pharmaceuticals.Phytother Res. 2015 Jan;29(1):1-13. doi: 10.1002/ptr.5222. Epub 2014 Sep 24. Phytother Res. 2015. PMID: 25256791 Review.
-
A Systematic and Mechanistic Review on the Phytopharmacological Properties of Alhagi Species.Anc Sci Life. 2016 Oct-Dec;36(2):65-71. doi: 10.4103/asl.ASL_37_16. Anc Sci Life. 2016. PMID: 28446826 Free PMC article. Review.
Cited by
-
Aquificae overcomes competition by archaeal thermophiles, and crowding by bacterial mesophiles, to dominate the boiling vent-water of a Trans-Himalayan sulfur-borax spring.PLoS One. 2024 Oct 25;19(10):e0310595. doi: 10.1371/journal.pone.0310595. eCollection 2024. PLoS One. 2024. PMID: 39453910 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

