Impact of the number of mutations in survival and response outcomes to hypomethylating agents in patients with myelodysplastic syndromes or myelodysplastic/myeloproliferative neoplasms
- PMID: 29515765
- PMCID: PMC5839396
- DOI: 10.18632/oncotarget.23882
Impact of the number of mutations in survival and response outcomes to hypomethylating agents in patients with myelodysplastic syndromes or myelodysplastic/myeloproliferative neoplasms
Abstract
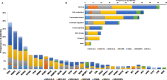
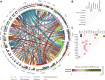
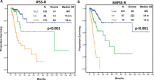
The prognostic and predictive value of sequencing analysis in myelodysplastic syndromes (MDS) has not been fully integrated into clinical practice. We performed whole exome sequencing (WES) of bone marrow samples from 83 patients with MDS and 31 with MDS/MPN identifying 218 driver mutations in 31 genes in 98 (86%) patients. A total of 65 (57%) patients received therapy with hypomethylating agents. By univariate analysis, mutations in BCOR, STAG2, TP53 and SF3B1 significantly influenced survival. Increased number of mutations (≥ 3), but not clonal heterogeneity, predicted for shorter survival and LFS. Presence of 3 or more mutations also predicted for lower likelihood of response (26 vs 50%, p = 0.055), and shorter response duration (3.6 vs 26.5 months, p = 0.022). By multivariate analysis, TP53 mutations (HR 3.1, CI 1.3-7.5, p = 0.011) and number of mutations (≥ 3) (HR 2.5, CI 1.3-4.8, p = 0.005) predicted for shorter survival. A novel prognostic model integrating this mutation data with IPSS-R separated patients into three categories with median survival of not reached, 29 months and 12 months respectively (p < 0.001) and increased stratification potential, compared to IPSS-R, in patients with high/very-high IPSS-R. This model was validated in a separate cohort of 413 patients with untreated MDS. Although the use of WES did not provide significant more information than that obtained with targeted sequencing, our findings indicate that increased number of mutations is an independent prognostic factor in MDS and that mutation data can add value to clinical prognostic models.
Keywords: chronic myelomonocytic leukemia; mutations; myelodysplastic syndromes; prognosis; response.
Conflict of interest statement
CONFLICTS OF INTEREST The authors do not disclose any conflicts of interest.
Figures


References
-
- Greenberg P, Cox C, LeBeau MM, Fenaux P, Morel P, Sanz G, Sanz M, Vallespi T, Hamblin T, Oscier D, Ohyashiki K, Toyama K, Aul C, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89:2079–88. - PubMed
-
- Greenberg PL, Tuechler H, Schanz J, Sanz G, Garcia-Manero G, Sole F, Bennett JM, Bowen D, Fenaux P, Dreyfus F, Kantarjian H, Kuendgen A, Levis A, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120:2454–65. https://doi.org/10.1182/blood-2012-03-420489. - DOI - PMC - PubMed
-
- Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G, Kantarjian H, Raza A, Levine RL, Neuberg D, Ebert BL. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364:2496–506. https://doi.org/10.1056/NEJMoa1013343. - DOI - PMC - PubMed
-
- Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, Schnittger S, Sanada M, Kon A, Alpermann T, Yoshida K, Roller A, Nadarajah N, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–7. https://doi.org/10.1038/leu.2013.336. - DOI - PMC - PubMed
-
- Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, Yoon CJ, Ellis P, Wedge DC, Pellagatti A, Shlien A, Groves MJ, Forbes SA, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–27. quiz 3699. https://doi.org/10.1182/blood-2013-08-518886. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

