Identification of allosteric disulfides from labile bonds in X-ray structures
- PMID: 29515832
- PMCID: PMC5830721
- DOI: 10.1098/rsos.171058
Identification of allosteric disulfides from labile bonds in X-ray structures
Abstract
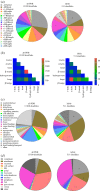
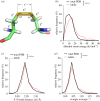
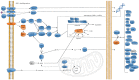
Protein disulfide bonds link pairs of cysteine sulfur atoms and are either structural or functional motifs. The allosteric disulfides control the function of the protein in which they reside when cleaved or formed. Here, we identify potential allosteric disulfides in all Protein Data Bank X-ray structures from bonds that are present in some molecules of a protein crystal but absent in others, or present in some structures of a protein but absent in others. We reasoned that the labile nature of these disulfides signifies a propensity for cleavage and so possible allosteric regulation of the protein in which the bond resides. A total of 511 labile disulfide bonds were identified. The labile disulfides are more stressed than the average bond, being characterized by high average torsional strain and stretching of the sulfur-sulfur bond and neighbouring bond angles. This pre-stress likely underpins their susceptibility to cleavage. The coagulation, complement and oxygen-sensing hypoxia inducible factor-1 pathways, which are known or have been suggested to be regulated by allosteric disulfides, are enriched in proteins containing labile disulfides. The identification of labile disulfide bonds will facilitate the study of this post-translational modification.
Keywords: allosteric disulfides; coagulation; complement; oxygen-sensing; redox.
Conflict of interest statement
We declare we have no competing interests.
Figures



Similar articles
-
Identification of allosteric disulfides from prestress analysis.Biophys J. 2014 Aug 5;107(3):672-681. doi: 10.1016/j.bpj.2014.06.025. Biophys J. 2014. PMID: 25099806 Free PMC article.
-
Control of mature protein function by allosteric disulfide bonds.Antioxid Redox Signal. 2011 Jan 1;14(1):113-26. doi: 10.1089/ars.2010.3620. Epub 2010 Oct 28. Antioxid Redox Signal. 2011. PMID: 20831445 Review.
-
Allosteric disulfide bonds.Biochemistry. 2006 Jun 20;45(24):7429-33. doi: 10.1021/bi0603064. Biochemistry. 2006. PMID: 16768438
-
Post-translational control of protein function by disulfide bond cleavage.Antioxid Redox Signal. 2013 May 20;18(15):1987-2015. doi: 10.1089/ars.2012.4807. Epub 2013 Feb 15. Antioxid Redox Signal. 2013. PMID: 23198756 Review.
-
Search for allosteric disulfide bonds in NMR structures.BMC Struct Biol. 2007 Jul 20;7:49. doi: 10.1186/1472-6807-7-49. BMC Struct Biol. 2007. PMID: 17640393 Free PMC article.
Cited by
-
Scientific review of the aesthetic uses of botulinum toxin type A.Arch Craniofac Surg. 2021 Feb;22(1):1-10. doi: 10.7181/acfs.2021.00003. Epub 2021 Feb 20. Arch Craniofac Surg. 2021. PMID: 33714246 Free PMC article.
-
Antioxidative Potency of Dolphin Serum Albumin Is Stronger Than That of Human Serum Albumin Irrespective of Substitution of 34Cysteine With Serine.Front Physiol. 2020 Nov 2;11:598451. doi: 10.3389/fphys.2020.598451. eCollection 2020. Front Physiol. 2020. PMID: 33224041 Free PMC article.
-
A redox-active switch in fructosamine-3-kinases expands the regulatory repertoire of the protein kinase superfamily.Sci Signal. 2020 Jul 7;13(639):eaax6313. doi: 10.1126/scisignal.aax6313. Sci Signal. 2020. PMID: 32636308 Free PMC article.
-
Cys.sqlite: A Structured-Information Approach to the Comprehensive Analysis of Cysteine Disulfide Bonds in the Protein Databank.J Chem Inf Model. 2019 Feb 25;59(2):931-943. doi: 10.1021/acs.jcim.8b00950. Epub 2019 Feb 15. J Chem Inf Model. 2019. PMID: 30694665 Free PMC article.
-
Structure of Usutu virus SAAR-1776 displays fusion loop asymmetry.Proc Natl Acad Sci U S A. 2021 Aug 24;118(34):e2107408118. doi: 10.1073/pnas.2107408118. Proc Natl Acad Sci U S A. 2021. PMID: 34417300 Free PMC article.
References
-
- Cook KM, Hogg PJ. 2013. Post-translational control of protein function by disulfide bond cleavage. Antioxid. Redox Signal. 18, 1987–2015. (doi:10.1089/ars.2012.4807) - DOI - PubMed
-
- Butera D, Cook KM, Chiu J, Wong JW, Hogg PJ. 2014. Control of blood proteins by functional disulfide bonds. Blood 123, 2000–2007. (doi:10.1182/blood-2014-01-549816) - DOI - PMC - PubMed
-
- Hogg PJ. 2013. Targeting allosteric disulphide bonds in cancer. Nat. Rev. Cancer 13, 425–431. (doi:10.1038/nrc3519) - DOI - PubMed
-
- Wouters MA, Lau KK, Hogg PJ. 2004. Cross-strand disulphides in cell entry proteins: poised to act. Bioessays 26, 73–79. (doi:10.1002/bies.10413) - DOI - PubMed
-
- Bekendam RH, et al. 2016. A substrate-driven allosteric switch that enhances PDI catalytic activity. Nat. Commun. 7, 12579 (doi:10.1038/ncomms12579) - DOI - PMC - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

