Unique mechanisms of connective tissue growth factor regulation in airway smooth muscle in asthma: Relationship with airway remodelling
- PMID: 29516637
- PMCID: PMC5908101
- DOI: 10.1111/jcmm.13576
Unique mechanisms of connective tissue growth factor regulation in airway smooth muscle in asthma: Relationship with airway remodelling
Abstract
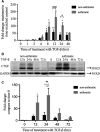
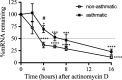
Neovascularization, increased basal membrane thickness and increased airway smooth muscle (ASM) bulk are hallmarks of airway remodelling in asthma. In this study, we examined connective tissue growth factor (CTGF) dysregulation in human lung tissue and animal models of allergic airway disease. Immunohistochemistry revealed that ASM cells from patients with severe asthma (A) exhibited high expression of CTGF, compared to mild and non-asthmatic (NA) tissues. This finding was replicated in a sheep model of allergic airways disease. In vitro, transforming growth factor (TGF)-β increased CTGF expression both in NA- and A-ASM cells but the expression was higher in A-ASM at both the mRNA and protein level as assessed by PCR and Western blot. Transfection of CTGF promoter-luciferase reporter constructs into NA- and A-ASM cells indicated that no region of the CTGF promoter (-1500 to +200 bp) displayed enhanced activity in the presence of TGF-β. However, in silico analysis of the CTGF promoter suggested that distant transcription factor binding sites may influence CTGF promoter activation by TGF-β in ASM cells. The discord between promoter activity and mRNA expression was also explained, in part, by differential post-transcriptional regulation in A-ASM cells due to enhanced mRNA stability for CTGF. In patients, higher CTGF gene expression in bronchial biopsies was correlated with increased basement membrane thickness indicating that the enhanced CTGF expression in A-ASM may contribute to airway remodelling in asthma.
Keywords: airway remodelling; airway smooth muscle; asthma; connective tissue growth factor.
© 2018 The Authors. Journal of Cellular and Molecular Medicine published by John Wiley & Sons Ltd and Foundation for Cellular and Molecular Medicine.
Figures

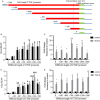
 ) were placed upstream of a Luciferase reporter construct (
) were placed upstream of a Luciferase reporter construct ( ). Secretion of alkaline phosphatase (
). Secretion of alkaline phosphatase (

Similar articles
-
STAT3 potentiates the ability of airway smooth muscle cells to promote angiogenesis by regulating VEGF signalling.Exp Physiol. 2017 May 1;102(5):598-606. doi: 10.1113/EP086136. Epub 2017 Apr 7. Exp Physiol. 2017. PMID: 28295786
-
Human airway smooth muscle cell proliferation from asthmatics is negatively regulated by semaphorin3A.Oncotarget. 2016 Dec 6;7(49):80238-80251. doi: 10.18632/oncotarget.12884. Oncotarget. 2016. PMID: 27791986 Free PMC article.
-
Differential estrogen-receptor activation regulates extracellular matrix deposition in human airway smooth muscle remodeling via NF-κB pathway.FASEB J. 2019 Dec;33(12):13935-13950. doi: 10.1096/fj.201901340R. Epub 2019 Oct 22. FASEB J. 2019. PMID: 31638834 Free PMC article.
-
Connective tissue growth factor: a role in airway remodelling in asthma?Clin Exp Pharmacol Physiol. 2005 Nov;32(11):988-94. doi: 10.1111/j.1440-1681.2005.04296.x. Clin Exp Pharmacol Physiol. 2005. PMID: 16405457 Review.
-
Transforming growth factor-beta1 in asthmatic airway smooth muscle enlargement: is fibroblast growth factor-2 required?Clin Exp Allergy. 2010 May;40(5):710-24. doi: 10.1111/j.1365-2222.2010.03497.x. Clin Exp Allergy. 2010. PMID: 20447083 Review.
Cited by
-
Cholinergic neuroplasticity in asthma driven by TrkB signaling.FASEB J. 2020 Jun;34(6):7703-7717. doi: 10.1096/fj.202000170R. Epub 2020 Apr 11. FASEB J. 2020. PMID: 32277855 Free PMC article.
-
Quantitative relationships between SMAD dynamics and target gene activation kinetics in single live cells.Sci Rep. 2019 Mar 29;9(1):5372. doi: 10.1038/s41598-019-41870-2. Sci Rep. 2019. PMID: 30926874 Free PMC article.
-
Effect of curcumin nanoparticles on proliferation and migration of mouse airway smooth muscle cells and airway inflammatory infiltration.Front Pharmacol. 2024 Apr 19;15:1344333. doi: 10.3389/fphar.2024.1344333. eCollection 2024. Front Pharmacol. 2024. PMID: 38708080 Free PMC article.
-
Bronchial thermoplasty in severe asthma: a real-world study on efficacy and gene profiling.Allergy Asthma Clin Immunol. 2022 May 9;18(1):39. doi: 10.1186/s13223-022-00680-4. Allergy Asthma Clin Immunol. 2022. PMID: 35534846 Free PMC article.
-
Fibroblast-to-myofibroblast transition in bronchial asthma.Cell Mol Life Sci. 2018 Nov;75(21):3943-3961. doi: 10.1007/s00018-018-2899-4. Epub 2018 Aug 12. Cell Mol Life Sci. 2018. PMID: 30101406 Free PMC article. Review.
References
-
- Asthma GIf . 2016 GINA Report, Global strategy for Asthma Management and Prevention; 2016.
-
- Burgess JK. The extracellular matrix: friend or foe in airway disease? Minerva Pneumol. 2010;49:219‐236.
-
- James AL, Elliot JG, Jones RL, et al. Airway smooth muscle hypertrophy and hyperplasia in asthma. Am J Respir Crit Care Med. 2012;185:1058‐1064. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

