Evolution and Stress Responses of Gossypium hirsutum SWEET Genes
- PMID: 29517986
- PMCID: PMC5877630
- DOI: 10.3390/ijms19030769
Evolution and Stress Responses of Gossypium hirsutum SWEET Genes
Abstract
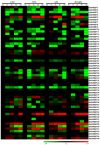
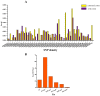
The SWEET (sugars will eventually be exported transporters) proteins are sugar efflux transporters containing the MtN3_saliva domain, which affects plant development as well as responses to biotic and abiotic stresses. These proteins have not been functionally characterized in the tetraploid cotton, Gossypium hirsutum, which is a widely cultivated cotton species. In this study, we comprehensively analyzed the cotton SWEET gene family. A total of 55 putative G. hirsutumSWEET genes were identified. The GhSWEET genes were classified into four clades based on a phylogenetic analysis and on the examination of gene structural features. Moreover, chromosomal localization and an analysis of homologous genes in Gossypium arboreum, Gossypium raimondii, and G. hirsutum suggested that a whole-genome duplication, several tandem duplications, and a polyploidy event contributed to the expansion of the cotton SWEET gene family, especially in Clade III and IV. Analyses of cis-acting regulatory elements in the promoter regions, expression profiles, and artificial selection revealed that the GhSWEET genes were likely involved in cotton developmental processes and responses to diverse stresses. These findings may clarify the evolution of G. hirsutum SWEET gene family and may provide a foundation for future functional studies of SWEET proteins regarding cotton development and responses to abiotic stresses.
Keywords: SWEET; artificial selection; cotton; evolution; stress response.
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures






Similar articles
-
Genome-wide characterization and phylogenetic analysis of GSK gene family in three species of cotton: evidence for a role of some GSKs in fiber development and responses to stress.BMC Plant Biol. 2018 Dec 4;18(1):330. doi: 10.1186/s12870-018-1526-8. BMC Plant Biol. 2018. PMID: 30514299 Free PMC article.
-
Expansion and stress responses of the AP2/EREBP superfamily in cotton.BMC Genomics. 2017 Jan 31;18(1):118. doi: 10.1186/s12864-017-3517-9. BMC Genomics. 2017. PMID: 28143399 Free PMC article.
-
Genome-wide comparative analysis of NBS-encoding genes in four Gossypium species.BMC Genomics. 2017 Apr 12;18(1):292. doi: 10.1186/s12864-017-3682-x. BMC Genomics. 2017. PMID: 28403834 Free PMC article.
-
Genome-Wide Characterization of the ABI3 Gene Family in Cotton.Genes (Basel). 2025 Jul 23;16(8):854. doi: 10.3390/genes16080854. Genes (Basel). 2025. PMID: 40869902 Free PMC article.
-
Genome-wide identification of lipoxygenase gene family in cotton and functional characterization in response to abiotic stresses.BMC Genomics. 2018 Aug 9;19(1):599. doi: 10.1186/s12864-018-4985-2. BMC Genomics. 2018. PMID: 30092779 Free PMC article.
Cited by
-
SWEET Gene Family in Medicago truncatula: Genome-Wide Identification, Expression and Substrate Specificity Analysis.Plants (Basel). 2019 Sep 9;8(9):338. doi: 10.3390/plants8090338. Plants (Basel). 2019. PMID: 31505820 Free PMC article.
-
Cloning and Functional Assessments of Floral-Expressed SWEET Transporter Genes from Jasminum sambac.Int J Mol Sci. 2019 Aug 16;20(16):4001. doi: 10.3390/ijms20164001. Int J Mol Sci. 2019. PMID: 31426432 Free PMC article.
-
Molecular Evolution and Stress and Phytohormone Responsiveness of SUT Genes in Gossypium hirsutum.Front Genet. 2018 Oct 23;9:494. doi: 10.3389/fgene.2018.00494. eCollection 2018. Front Genet. 2018. PMID: 30405700 Free PMC article.
-
SWEET Transporters and the Potential Functions of These Sequences in Tea (Camellia sinensis).Front Genet. 2021 Mar 31;12:655843. doi: 10.3389/fgene.2021.655843. eCollection 2021. Front Genet. 2021. PMID: 33868386 Free PMC article.
-
Evolutionary Relationships and Divergence of KNOTTED1-Like Family Genes Involved in Salt Tolerance and Development in Cotton (Gossypium hirsutum L.).Front Plant Sci. 2021 Dec 14;12:774161. doi: 10.3389/fpls.2021.774161. eCollection 2021. Front Plant Sci. 2021. PMID: 34970288 Free PMC article.
References
-
- Srivastava A.C., Ganesan S., Ismail I.O., Ayre B.G. Functional characterization of the Arabidopsis AtSUC2 Sucrose/H+ symporter by tissue-specific complementation reveals an essential role in phloem loading but not in long-distance transport. Plant Physiol. 2008;148:200–211. doi: 10.1104/pp.108.124776. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

