Emerging Roles of p53 Family Members in Glucose Metabolism
- PMID: 29518025
- PMCID: PMC5877637
- DOI: 10.3390/ijms19030776
Emerging Roles of p53 Family Members in Glucose Metabolism
Abstract
Glucose is the key source for most organisms to provide energy, as well as the key source for metabolites to generate building blocks in cells. The deregulation of glucose homeostasis occurs in various diseases, including the enhanced aerobic glycolysis that is observed in cancers, and insulin resistance in diabetes. Although p53 is thought to suppress tumorigenesis primarily by inducing cell cycle arrest, apoptosis, and senescence in response to stress, the non-canonical functions of p53 in cellular energy homeostasis and metabolism are also emerging as critical factors for tumor suppression. Increasing evidence suggests that p53 plays a significant role in regulating glucose homeostasis. Furthermore, the p53 family members p63 and p73, as well as gain-of-function p53 mutants, are also involved in glucose metabolism. Indeed, how this protein family regulates cellular energy levels is complicated and difficult to disentangle. This review discusses the roles of the p53 family in multiple metabolic processes, such as glycolysis, gluconeogenesis, aerobic respiration, and autophagy. We also discuss how the dysregulation of the p53 family in these processes leads to diseases such as cancer and diabetes. Elucidating the complexities of the p53 family members in glucose homeostasis will improve our understanding of these diseases.
Keywords: autophagy; cancer; diabetes; glucose metabolism; glycolysis; mitochondria; p53; p53 mutant; p63; p73.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

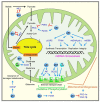
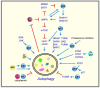
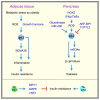
References
-
- Brady C.A., Jiang D., Mello S.S., Johnson T.M., Jarvis L.A., Kozak M.M., Broz D.K., Basak S., Park E.J., McLaughlin M.E., et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145:571–583. doi: 10.1016/j.cell.2011.03.035. - DOI - PMC - PubMed
-
- Valente L.J., Gray D.H., Michalak E.M., Pinon-Hofbauer J., Egle A., Scott C.L., Janic A., Strasser A. p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa. Cell Rep. 2013;3:1339–1345. doi: 10.1016/j.celrep.2013.04.012. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

