Analysis of Gene Regulatory Networks of Maize in Response to Nitrogen
- PMID: 29518046
- PMCID: PMC5867872
- DOI: 10.3390/genes9030151
Analysis of Gene Regulatory Networks of Maize in Response to Nitrogen
Abstract
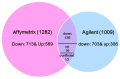
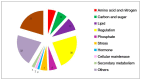
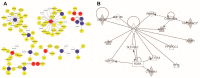
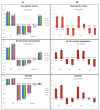
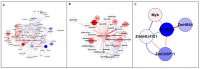
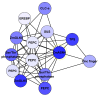
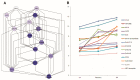
Nitrogen (N) fertilizer has a major influence on the yield and quality. Understanding and optimising the response of crop plants to nitrogen fertilizer usage is of central importance in enhancing food security and agricultural sustainability. In this study, the analysis of gene regulatory networks reveals multiple genes and biological processes in response to N. Two microarray studies have been used to infer components of the nitrogen-response network. Since they used different array technologies, a map linking the two probe sets to the maize B73 reference genome has been generated to allow comparison. Putative Arabidopsis homologues of maize genes were used to query the Biological General Repository for Interaction Datasets (BioGRID) network, which yielded the potential involvement of three transcription factors (TFs) (GLK5, MADS64 and bZIP108) and a Calcium-dependent protein kinase. An Artificial Neural Network was used to identify influential genes and retrieved bZIP108 and WRKY36 as significant TFs in both microarray studies, along with genes for Asparagine Synthetase, a dual-specific protein kinase and a protein phosphatase. The output from one study also suggested roles for microRNA (miRNA) 399b and Nin-like Protein 15 (NLP15). Co-expression-network analysis of TFs with closely related profiles to known Nitrate-responsive genes identified GLK5, GLK8 and NLP15 as candidate regulators of genes repressed under low Nitrogen conditions, while bZIP108 might play a role in gene activation.
Keywords: maize; network analysis; nitrogen response genes; regulatory network inference.
Conflict of interest statement
The authors declare no conflicts of interest. The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures


Similar articles
-
Cross-Species Network Analysis Uncovers Conserved Nitrogen-Regulated Network Modules in Rice.Plant Physiol. 2015 Aug;168(4):1830-43. doi: 10.1104/pp.114.255877. Epub 2015 Jun 4. Plant Physiol. 2015. PMID: 26045464 Free PMC article.
-
Maize network analysis revealed gene modules involved in development, nutrients utilization, metabolism, and stress response.BMC Plant Biol. 2017 Aug 1;17(1):131. doi: 10.1186/s12870-017-1077-4. BMC Plant Biol. 2017. PMID: 28764653 Free PMC article.
-
Transcriptomic analysis highlights reciprocal interactions of urea and nitrate for nitrogen acquisition by maize roots.Plant Cell Physiol. 2015 Mar;56(3):532-48. doi: 10.1093/pcp/pcu202. Epub 2014 Dec 17. Plant Cell Physiol. 2015. PMID: 25524070
-
Discovery of core biotic stress responsive genes in Arabidopsis by weighted gene co-expression network analysis.PLoS One. 2015 Mar 2;10(3):e0118731. doi: 10.1371/journal.pone.0118731. eCollection 2015. PLoS One. 2015. PMID: 25730421 Free PMC article.
-
Regulatory modules controlling early shade avoidance response in maize seedlings.BMC Genomics. 2016 Mar 31;17:269. doi: 10.1186/s12864-016-2593-6. BMC Genomics. 2016. PMID: 27030359 Free PMC article.
Cited by
-
Low nitrogen availability inhibits the phosphorus starvation response in maize (Zea mays ssp. mays L.).BMC Plant Biol. 2021 Jun 5;21(1):259. doi: 10.1186/s12870-021-02997-5. BMC Plant Biol. 2021. PMID: 34090337 Free PMC article.
-
Analysis of Gene Regulatory Networks of Taro (Colocasia esculenta (L.) Schott.) Soluble Starch Synthase Based on DeGN and KASP Marker Development.Int J Genomics. 2025 Mar 1;2025:9953367. doi: 10.1155/ijog/9953367. eCollection 2025. Int J Genomics. 2025. PMID: 40226354 Free PMC article.
-
Agro-physiological and transcriptome profiling reveal key genes associated with potato tuberization under different nitrogen regimes in aeroponics.PLoS One. 2025 Mar 28;20(3):e0320313. doi: 10.1371/journal.pone.0320313. eCollection 2025. PLoS One. 2025. PMID: 40153446 Free PMC article.
-
Insights on Phytohormonal Crosstalk in Plant Response to Nitrogen Stress: A Focus on Plant Root Growth and Development.Int J Mol Sci. 2023 Feb 11;24(4):3631. doi: 10.3390/ijms24043631. Int J Mol Sci. 2023. PMID: 36835044 Free PMC article. Review.
-
Large Scale Proteomic Data and Network-Based Systems Biology Approaches to Explore the Plant World.Proteomes. 2018 Jun 3;6(2):27. doi: 10.3390/proteomes6020027. Proteomes. 2018. PMID: 29865292 Free PMC article.
References
-
- Goulding K. Nitrate leaching from arable and horticultural land. Soil Use Manag. 2000;16:145–151. doi: 10.1111/j.1475-2743.2000.tb00218.x. - DOI
-
- Agrama H.A.S., Zakaria A.G., Said F.B., Tuinstra M. Identification of quantitative trait loci for nitrogen use efficiency in maize. Mol. Breed. 1999;5:187–195. doi: 10.1023/A:1009669507144. - DOI
-
- Bertin P., Gallais A. Genetic variation for nitrogen use efficiency in a set of recombinant maize inbred lines. I. Agrophysiological results. Maydica. 2000;45:53–66.
-
- Hirel B., Bertin P., Quillere I., Bourdoncle W., Attagnant C., Dellay C., Gouy A., Cadiou S., Retailliau C., Falque M., et al. Towards a better understanding of the genetic and physiological basis for nitrogen use efficiency in maize. Plant Physiol. 2001;125:1258–1270. doi: 10.1104/pp.125.3.1258. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

