Potential Roles of Dec and Bmal1 Genes in Interconnecting Circadian Clock and Energy Metabolism
- PMID: 29518061
- PMCID: PMC5877642
- DOI: 10.3390/ijms19030781
Potential Roles of Dec and Bmal1 Genes in Interconnecting Circadian Clock and Energy Metabolism
Abstract
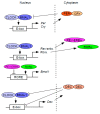
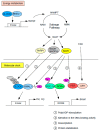
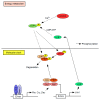
The daily rhythm of mammalian energy metabolism is subject to the circadian clock system, which is made up of the molecular clock machinery residing in nearly all cells throughout the body. The clock genes have been revealed not only to form the molecular clock but also to function as a mediator that regulates both circadian and metabolic functions. While the circadian signals generated by clock genes produce metabolic rhythms, clock gene function is tightly coupled to fundamental metabolic processes such as glucose and lipid metabolism. Therefore, defects in the clock genes not only result in the dysregulation of physiological rhythms but also induce metabolic disorders including diabetes and obesity. Among the clock genes, Dec1 (Bhlhe40/Stra13/Sharp2), Dec2 (Bhlhe41/Sharp1), and Bmal1 (Mop3/Arntl) have been shown to be particularly relevant to the regulation of energy metabolism at the cellular, tissue, and organismal levels. This paper reviews our current knowledge of the roles of Dec1, Dec2, and Bmal1 in coordinating the circadian and metabolic pathways.
Keywords: Bmal1; Dec1; Dec2; clock gene; energy metabolism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

