Quantitative Proteomic Approach Targeted to Fibrinogen β Chain in Tissue Gastric Carcinoma
- PMID: 29518939
- PMCID: PMC5877620
- DOI: 10.3390/ijms19030759
Quantitative Proteomic Approach Targeted to Fibrinogen β Chain in Tissue Gastric Carcinoma
Abstract
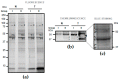
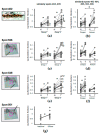
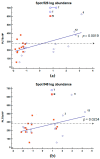
Elevated plasma fibrinogen levels and tumor progression in patients with gastric cancer (GC) have been largely reported. However, distinct fibrinogen chains and domains have different effects on coagulation, inflammation, and angiogenesis. The aim of this study was to characterize fibrinogen β chain (FGB) in GC tissues. Retrospectively we analyzed the data of matched pairs of normal (N) and malignant tissues (T) of 28 consecutive patients with GC at diagnosis by combining one- and two-dimensional electrophoresis (1DE and 2DE) with immunoblotting and mass spectrometry together with two-dimensional difference in gel electrophoresis (2D-DIGE). 1DE showed bands of the intact FGB at 50 kDa and the cleaved forms containing the fragment D at ~37-40 kDa, which corresponded to 19 spots in 2DE. In particular, spot 402 at ~50 kDa and spots 526 and 548 at ~37 kDa were of interest by showing an increased expression in tumor tissues. A higher content of spot 402 was associated with stomach antrum, while spots 526 and 548 amounts correlated with corpus and high platelet count (>208 × 10⁸/L). The quantification of FGB and cleaved products may help to further characterize the interconnections between GC and platelet/coagulation pathways.
Keywords: DIGE; FGB; biomarker; coagulation; comparative proteomics; fibrinogen β chain; gastric cancer; platelets.
Conflict of interest statement
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures




Similar articles
-
Proteomic analysis in clear cell renal cell carcinoma: identification of differentially expressed protein by 2-D DIGE.Mol Biosyst. 2012 Apr;8(4):1040-51. doi: 10.1039/c2mb05390j. Epub 2012 Feb 8. Mol Biosyst. 2012. PMID: 22315040
-
[Application of proteomics in the research of discrepancy proteins in gastric cancer].Zhonghua Wei Chang Wai Ke Za Zhi. 2006 Nov;9(6):534-7. Zhonghua Wei Chang Wai Ke Za Zhi. 2006. PMID: 17143804 Chinese.
-
Proteomics-based identification of a group of apoptosis-related proteins and biomarkers in gastric cancer.Int J Oncol. 2011 Feb;38(2):375-83. doi: 10.3892/ijo.2010.873. Epub 2010 Dec 15. Int J Oncol. 2011. PMID: 21165559
-
Mass Spectrometry-Based Protein Quantification.Adv Exp Med Biol. 2016;919:255-279. doi: 10.1007/978-3-319-41448-5_15. Adv Exp Med Biol. 2016. PMID: 27975224 Review.
-
Differential proteomics mass spectrometry of melanosis coli.Am J Transl Res. 2020 Jul 15;12(7):3133-3148. eCollection 2020. Am J Transl Res. 2020. PMID: 32774690 Free PMC article. Review.
Cited by
-
Identification and Interaction Analysis of Molecular Markers in Pancreatic Ductal Adenocarcinoma by Bioinformatics and Next-Generation Sequencing Data Analysis.Bioinform Biol Insights. 2023 Jul 25;17:11779322231186719. doi: 10.1177/11779322231186719. eCollection 2023. Bioinform Biol Insights. 2023. PMID: 37529485 Free PMC article.
-
Early diagnosis of colorectal cancer via plasma proteomic analysis of CRC and advanced adenomatous polyp.Gastroenterol Hepatol Bed Bench. 2019 Fall;12(4):328-339. Gastroenterol Hepatol Bed Bench. 2019. PMID: 31749922 Free PMC article.
-
A machine learning-based approach to predicting the malignant and metastasis of thyroid cancer.Front Oncol. 2022 Dec 19;12:938292. doi: 10.3389/fonc.2022.938292. eCollection 2022. Front Oncol. 2022. PMID: 36601485 Free PMC article.
-
Intracellular peptides in SARS-CoV-2-infected patients.iScience. 2023 Aug 6;26(9):107542. doi: 10.1016/j.isci.2023.107542. eCollection 2023 Sep 15. iScience. 2023. PMID: 37636076 Free PMC article.
-
Label-based comparative proteomics of oral mucosal tissue to understand progression of precancerous lesions to oral squamous cell carcinoma.Biochem Biophys Rep. 2024 Oct 13;40:101842. doi: 10.1016/j.bbrep.2024.101842. eCollection 2024 Dec. Biochem Biophys Rep. 2024. PMID: 39483176 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

