Unravelling the Effects of the Mutation m.3571insC/MT-ND1 on Respiratory Complexes Structural Organization
- PMID: 29518970
- PMCID: PMC5877625
- DOI: 10.3390/ijms19030764
Unravelling the Effects of the Mutation m.3571insC/MT-ND1 on Respiratory Complexes Structural Organization
Abstract
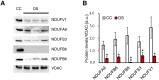
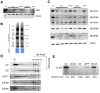
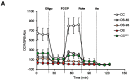
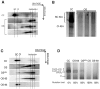
Mammalian respiratory complex I (CI) biogenesis requires both nuclear and mitochondria-encoded proteins and is mostly organized in respiratory supercomplexes. Among the CI proteins encoded by the mitochondrial DNA, NADH-ubiquinone oxidoreductase chain 1 (ND1) is a core subunit, evolutionary conserved from bacteria to mammals. Recently, ND1 has been recognized as a pivotal subunit in maintaining the structural and functional interaction among the hydrophilic and hydrophobic CI arms. A critical role of human ND1 both in CI biogenesis and in the dynamic organization of supercomplexes has been depicted, although the proof of concept is still missing and the critical amount of ND1 protein necessary for a proper assembly of both CI and supercomplexes is not defined. By exploiting a unique model in which human ND1 is allotopically re-expressed in cells lacking the endogenous protein, we demonstrated that the lack of this protein induces a stall in the multi-step process of CI biogenesis, as well as the alteration of supramolecular organization of respiratory complexes. We also defined a mutation threshold for the m.3571insC truncative mutation in mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 1 (MT-ND1), below which CI and its supramolecular organization is recovered, strengthening the notion that a certain amount of human ND1 is required for CI and supercomplexes biogenesis.
Keywords: MT-ND1; ND1; OXPHOS; mitochondria; mtDNA mutation; respirasome; respiratory complex I; respiratory complexes; supercomplexes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Mitochondrial complex I subunit MT-ND1 mutations affect disease progression.Heliyon. 2024 Mar 28;10(7):e28808. doi: 10.1016/j.heliyon.2024.e28808. eCollection 2024 Apr 15. Heliyon. 2024. PMID: 38596130 Free PMC article. Review.
-
Different mtDNA mutations modify tumor progression in dependence of the degree of respiratory complex I impairment.Hum Mol Genet. 2014 Mar 15;23(6):1453-66. doi: 10.1093/hmg/ddt533. Epub 2013 Oct 24. Hum Mol Genet. 2014. PMID: 24163135
-
Loss of mitochondrial DNA-encoded protein ND1 results in disruption of complex I biogenesis during early stages of assembly.FASEB J. 2016 Jun;30(6):2236-48. doi: 10.1096/fj.201500137R. Epub 2016 Feb 29. FASEB J. 2016. PMID: 26929434
-
Biochemical consequences of two clinically relevant ND-gene mutations in Escherichia coli respiratory complex I.Sci Rep. 2021 Jun 16;11(1):12641. doi: 10.1038/s41598-021-91631-3. Sci Rep. 2021. PMID: 34135385 Free PMC article.
-
Mitochondrial complex I deficiency of nuclear origin I. Structural genes.Mol Genet Metab. 2012 Feb;105(2):163-72. doi: 10.1016/j.ymgme.2011.11.188. Epub 2011 Nov 18. Mol Genet Metab. 2012. PMID: 22142868 Review.
Cited by
-
The Mitochondrial Genome in Aging and Disease and the Future of Mitochondrial Therapeutics.Biomedicines. 2022 Feb 18;10(2):490. doi: 10.3390/biomedicines10020490. Biomedicines. 2022. PMID: 35203698 Free PMC article. Review.
-
Codon optimization is an essential parameter for the efficient allotopic expression of mtDNA genes.Redox Biol. 2020 Feb;30:101429. doi: 10.1016/j.redox.2020.101429. Epub 2020 Jan 11. Redox Biol. 2020. PMID: 31981894 Free PMC article.
-
Mitochondrial complex I subunit MT-ND1 mutations affect disease progression.Heliyon. 2024 Mar 28;10(7):e28808. doi: 10.1016/j.heliyon.2024.e28808. eCollection 2024 Apr 15. Heliyon. 2024. PMID: 38596130 Free PMC article. Review.
-
Transcriptional Changes after Enniatins A, A1, B and B1 Ingestion in Rat Stomach, Liver, Kidney and Lower Intestine.Foods. 2021 Jul 14;10(7):1630. doi: 10.3390/foods10071630. Foods. 2021. PMID: 34359500 Free PMC article.
-
NDUFS3 depletion permits complex I maturation and reveals TMEM126A/OPA7 as an assembly factor binding the ND4-module intermediate.Cell Rep. 2021 Apr 20;35(3):109002. doi: 10.1016/j.celrep.2021.109002. Cell Rep. 2021. PMID: 33882309 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

