Transcriptome Profiling Reveals the EanI/R Quorum Sensing Regulon in Pantoea Ananatis LMG 2665T
- PMID: 29518982
- PMCID: PMC5867869
- DOI: 10.3390/genes9030148
Transcriptome Profiling Reveals the EanI/R Quorum Sensing Regulon in Pantoea Ananatis LMG 2665T
Abstract
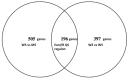
Pantoea ananatis LMG 2665T synthesizes and utilizes acyl homoserine lactones (AHLs) for signalling. The complete set of genes regulated by the EanI/R quorum sensing (QS) system in this strain is still not fully known. In this study, RNA-sequencing (RNA-seq) was used to identify the EanI/R regulon in LMG 2665T. Pairwise comparisons of LMG 2665T in the absence of AHLs (Optical density (OD)600 = 0.2) and in the presence of AHLs (OD600 = 0.5) were performed. Additionally, pairwise comparisons of LMG 2665T and its QS mutant at OD600 = 0.5 were undertaken. In total, 608 genes were differentially expressed between LMG 2665T at OD600 = 0.5 versus the same strain at OD600 = 0.2 and 701 genes were differentially expressed between LMG 2665T versus its QS mutant at OD600 = 0.5. A total of 196 genes were commonly differentially expressed between the two approaches. These constituted approximately 4.5% of the whole transcriptome under the experimental conditions used in this study. The RNA-seq data was validated by reverse transcriptase quantitative polymerase chain reaction (RT-qPCR). Genes found to be regulated by EanI/R QS were those coding for redox sensing, metabolism, flagella formation, flagella dependent motility, cell adhesion, biofilm formation, regulators, transport, chemotaxis, methyl accepting proteins, membrane proteins, cell wall synthesis, stress response and a large number of hypothetical proteins. The results of this study give insight into the genes that are regulated by the EanI/R system in LMG 2665T. Functional characterization of the QS regulated genes in LMG 2665T could assist in the formulation of control strategies for this plant pathogen.
Keywords: LMG 2665T; Pantoea ananatis; RNA-seq; acyl homoserine lactones; quorum sensing; regulon.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Functional Characterization of a Global Virulence Regulator Hfq and Identification of Hfq-Dependent sRNAs in the Plant Pathogen Pantoea ananatis.Front Microbiol. 2019 Sep 11;10:2075. doi: 10.3389/fmicb.2019.02075. eCollection 2019. Front Microbiol. 2019. PMID: 31572315 Free PMC article.
-
Characterization of two LuxI/R homologs in Pantoea ananatis LMG 2665T.Can J Microbiol. 2016 Nov;62(11):893-903. doi: 10.1139/cjm-2016-0143. Epub 2016 Jun 10. Can J Microbiol. 2016. PMID: 27510302
-
Transcriptome-based analysis of the Pantoea stewartii quorum-sensing regulon and identification of EsaR direct targets.Appl Environ Microbiol. 2014 Sep;80(18):5790-800. doi: 10.1128/AEM.01489-14. Epub 2014 Jul 11. Appl Environ Microbiol. 2014. PMID: 25015891 Free PMC article.
-
Bacterial quorum sensing in symbiotic and pathogenic relationships with hosts.Biosci Biotechnol Biochem. 2018 Mar;82(3):363-371. doi: 10.1080/09168451.2018.1433992. Epub 2018 Feb 9. Biosci Biotechnol Biochem. 2018. PMID: 29424268 Review.
-
Exploring the Function of Quorum Sensing Regulated Biofilms in Biological Wastewater Treatment: A Review.Int J Mol Sci. 2022 Aug 28;23(17):9751. doi: 10.3390/ijms23179751. Int J Mol Sci. 2022. PMID: 36077148 Free PMC article. Review.
Cited by
-
Transcriptional analysis reveals the metabolic state of Burkholderia zhejiangensis CEIB S4-3 during methyl parathion degradation.PeerJ. 2019 Apr 24;7:e6822. doi: 10.7717/peerj.6822. eCollection 2019. PeerJ. 2019. PMID: 31086743 Free PMC article.
-
Functional Characterization of a Global Virulence Regulator Hfq and Identification of Hfq-Dependent sRNAs in the Plant Pathogen Pantoea ananatis.Front Microbiol. 2019 Sep 11;10:2075. doi: 10.3389/fmicb.2019.02075. eCollection 2019. Front Microbiol. 2019. PMID: 31572315 Free PMC article.
References
-
- Cota L., Costa R., Silva D., Parreira D., Lana U., Casela C. First report of pathogenicity of Pantoea ananatis in sorghum (Sorghum bicolor) in Brazil. Australas. Plant Dis. Notes. 2010;5:120–122. doi: 10.1071/DN10044. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

