Proteasome Activity Is Influenced by the HECT_2 Protein Ipa1 in Budding Yeast
- PMID: 29519818
- PMCID: PMC5937183
- DOI: 10.1534/genetics.118.300744
Proteasome Activity Is Influenced by the HECT_2 Protein Ipa1 in Budding Yeast
Abstract
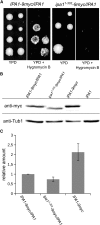
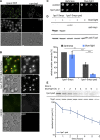
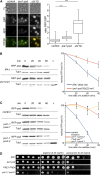
The ubiquitin-proteasome system (UPS) controls cellular functions by maintenance of a functional proteome and degradation of key regulatory proteins. Central to the UPS is the proteasome that adjusts the abundance of numerous proteins, thereby safeguarding their activity or initiating regulatory events. Here, we demonstrate that the essential Saccharomyces cerevisiae protein Yjr141w/Ipa1 (Important for cleavage and PolyAdenylation) belongs to the HECT_2 (homologous to E6-AP carboxyl terminus_2) family. We found that five cysteine residues within the HECT_2 family signature and the C-terminus are essential for Ipa1 activity. Furthermore, Ipa1 interacts with several ubiquitin-conjugating enzymes in vivo and localizes to the cytosol and nucleus. Importantly, Ipa1 has an impact on proteasome activity, which is indicated by the activation of the Rpn4 regulon as well as by decreased turnover of destabilized proteasome substrates in an IPA1 mutant. These changes in proteasome activity might be connected to reduced maturation or modification of proteasomal core particle proteins. Our results highlight the influence of Ipa1 on the UPS. The conservation within the HECT_2 family and the connection of the human HECT_2 family member to an age-related degeneration disease might suggest that HECT_2 family members share a conserved function linked to proteasome activity.
Keywords: Saccharomyces cerevisiae; polyadenylation and RNA cleavage; proteasome; protein degradation; ubiquitin–proteasome system.
Copyright © 2018 by the Genetics Society of America.
Figures





Similar articles
-
Regulation of the Ysh1 endonuclease of the mRNA cleavage/polyadenylation complex by ubiquitin-mediated degradation.RNA Biol. 2020 May;17(5):689-702. doi: 10.1080/15476286.2020.1724717. Epub 2020 Feb 12. RNA Biol. 2020. PMID: 32009536 Free PMC article.
-
Lysine Residues Are Not Required for Proteasome-Mediated Proteolysis of the Auxin/Indole Acidic Acid Protein IAA1.Plant Physiol. 2015 Jun;168(2):708-20. doi: 10.1104/pp.15.00402. Epub 2015 Apr 17. Plant Physiol. 2015. PMID: 25888615 Free PMC article.
-
Role of the ubiquitin-proteasome system in brain ischemia: friend or foe?Prog Neurobiol. 2014 Jan;112:50-69. doi: 10.1016/j.pneurobio.2013.10.003. Epub 2013 Oct 22. Prog Neurobiol. 2014. PMID: 24157661 Review.
-
Analyzing cell cycle-dependent degradation and ubiquitination in budding yeast.Methods Mol Biol. 2014;1170:343-56. doi: 10.1007/978-1-4939-0888-2_17. Methods Mol Biol. 2014. PMID: 24906322
-
The amazing ubiquitin-proteasome system: structural components and implication in aging.Int Rev Cell Mol Biol. 2015;314:171-237. doi: 10.1016/bs.ircmb.2014.09.002. Epub 2014 Nov 18. Int Rev Cell Mol Biol. 2015. PMID: 25619718 Review.
Cited by
-
The Protein Response of Salt-Tolerant Zygosaccharomyces rouxii to High-Temperature Stress during the Lag Phase.J Fungi (Basel). 2024 Jan 5;10(1):48. doi: 10.3390/jof10010048. J Fungi (Basel). 2024. PMID: 38248957 Free PMC article.
-
Regulation of the Ysh1 endonuclease of the mRNA cleavage/polyadenylation complex by ubiquitin-mediated degradation.RNA Biol. 2020 May;17(5):689-702. doi: 10.1080/15476286.2020.1724717. Epub 2020 Feb 12. RNA Biol. 2020. PMID: 32009536 Free PMC article.
-
Deficiency of UBE3D in mice leads to severe embryonic abnormalities and disrupts the mRNA of Homeobox genes via CPSF3.Cell Death Discov. 2025 Mar 12;11(1):99. doi: 10.1038/s41420-025-02387-y. Cell Death Discov. 2025. PMID: 40075082 Free PMC article.
-
Structure of the Yeast Cell Wall Integrity Sensor Wsc1 Reveals an Essential Role of Surface-Exposed Aromatic Clusters.J Fungi (Basel). 2022 Apr 8;8(4):379. doi: 10.3390/jof8040379. J Fungi (Basel). 2022. PMID: 35448610 Free PMC article.
References
-
- Alabrudzinska M., Skoneczny M., Skoneczna A., 2011. Diploid-specific [corrected] genome stability genes of S. cerevisiae: genomic screen reveals haploidization as an escape from persisting DNA rearrangement stress. PLoS One 6: e21124 (erratum: PLoS One 6) 10.1371/journal.pone.0021124 - DOI - PMC - PubMed
-
- Ausubel, F. M., R. E. Kingston, F. G. Seidman, K. Struhl, D. D. Moore et al. (Editors), 1995 Current Protocols in Molecular Biology. John Wiley and Sons, New York.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

