Analysis of Parent-of-Origin Effects on the X Chromosome in Asian and European Orofacial Cleft Triads Identifies Associations with DMD, FGF13, EGFL6, and Additional Loci at Xp22.2
- PMID: 29520293
- PMCID: PMC5827165
- DOI: 10.3389/fgene.2018.00025
Analysis of Parent-of-Origin Effects on the X Chromosome in Asian and European Orofacial Cleft Triads Identifies Associations with DMD, FGF13, EGFL6, and Additional Loci at Xp22.2
Abstract
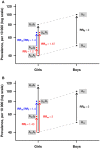
Background: Although both the mother's and father's alleles are present in the offspring, they may not operate at the same level. These parent-of-origin (PoO) effects have not yet been explored on the X chromosome, which motivated us to develop new methods for detecting such effects. Orofacial clefts (OFCs) exhibit sex-specific differences in prevalence and are examples of traits where a search for various types of effects on the X chromosome might be relevant. Materials and Methods: We upgraded our R-package Haplin to enable genome-wide analyses of PoO effects, as well as power simulations for different statistical models. 14,486 X-chromosome SNPs in 1,291 Asian and 1,118 European case-parent triads of isolated OFCs were available from a previous GWAS. For each ethnicity, cleft lip with or without cleft palate (CL/P) and cleft palate only (CPO) were analyzed separately using two X-inactivation models and a sliding-window approach to haplotype analysis. In addition, we performed analyses restricted to female offspring. Results: Associations were identified in "Dystrophin" (DMD, Xp21.2-p21.1), "Fibroblast growth factor 13" (FGF13, Xq26.3-q27.1) and "EGF-like domain multiple 6" (EGFL6, Xp22.2), with biologically plausible links to OFCs. Unlike EGFL6, the other associations on chromosomal region Xp22.2 had no apparent connections to OFCs. However, the Xp22.2 region itself is of potential interest because it contains genes for clefting syndromes [for example, "Oral-facial-digital syndrome 1" (OFD1) and "Midline 1" (MID1)]. Overall, the identified associations were highly specific for ethnicity, cleft subtype and X-inactivation model, except for DMD in which associations were identified in both CPO and CL/P, in the model with X-inactivation and in Europeans only. Discussion/Conclusion: The specificity of the associations for ethnicity, cleft subtype and X-inactivation model underscores the utility of conducting subanalyses, despite the ensuing need to adjust for additional multiple testing. Further investigations are needed to confirm the associations with DMD, EGF16, and FGF13. Furthermore, chromosomal region Xp22.2 appears to be a hotspot for genes implicated in clefting syndromes and thus constitutes an exciting direction to pursue in future OFCs research. More generally, the new methods presented here are readily adaptable to the study of X-linked PoO effects in other outcomes that use a family-based design.
Keywords: GWAS; Haplin; X chromosome; birth defects; case-parent triads; genetic epidemiology; orofacial clefts; parent-of-origin.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

