Genomic Characterisation of the Indigenous Irish Kerry Cattle Breed
- PMID: 29520297
- PMCID: PMC5827531
- DOI: 10.3389/fgene.2018.00051
Genomic Characterisation of the Indigenous Irish Kerry Cattle Breed
Abstract
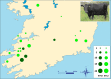
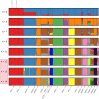
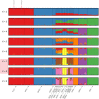
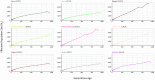
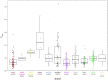
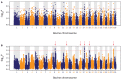
Kerry cattle are an endangered landrace heritage breed of cultural importance to Ireland. In the present study we have used genome-wide SNP array data to evaluate genomic diversity within the Kerry population and between Kerry cattle and other European breeds. Patterns of genetic differentiation and gene flow among breeds using phylogenetic trees with ancestry graphs highlighted historical gene flow from the British Shorthorn breed into the ancestral population of modern Kerry cattle. Principal component analysis (PCA) and genetic clustering emphasised the genetic distinctiveness of Kerry cattle relative to comparator British and European cattle breeds. Modelling of genetic effective population size (Ne) revealed a demographic trend of diminishing Ne over time and that recent estimated Ne values for the Kerry breed may be less than the threshold for sustainable genetic conservation. In addition, analysis of genome-wide autozygosity (FROH) showed that genomic inbreeding has increased significantly during the 20 years between 1992 and 2012. Finally, signatures of selection revealed genomic regions subject to natural and artificial selection as Kerry cattle adapted to the climate, physical geography and agro-ecology of southwest Ireland.
Keywords: cattle; conservation genomics; endangered breed; genetic diversity; inbreeding; population genomics; selection signature; single nucleotide polymorphism.
Figures




References
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

