Cholesterol enhances influenza binding avidity by controlling nanoscale receptor clustering
- PMID: 29520318
- PMCID: PMC5839467
- DOI: 10.1039/c7sc03236f
Cholesterol enhances influenza binding avidity by controlling nanoscale receptor clustering
Abstract
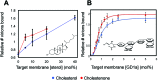
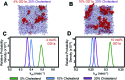
Influenza virus infects cells by binding to sialylated glycans on the cell surface. While the chemical structure of these glycans determines hemagglutinin-glycan binding affinity, bimolecular affinities are weak, so binding is avidity-dominated and driven by multivalent interactions. Here, we show that membrane spatial organization can control viral binding. Using single-virus fluorescence microscopy, we demonstrate that the sterol composition of the target membrane enhances viral binding avidity in a dose-dependent manner. Binding shows a cooperative dependence on concentration of receptors for influenza virus, as would be expected for a multivalent interaction. Surprisingly, the ability of sterols to promote viral binding is independent of their ability to support liquid-liquid phase separation in model systems. We develop a molecular explanation for this observation via molecular dynamics simulations, where we find that cholesterol promotes small-scale clusters of glycosphingolipid receptors. We propose a model whereby cholesterol orders the monomeric state of glycosphingolipid receptors, reducing the entropic penalty of receptor association and thus favoring multimeric complexes without phase separation. This model explains how cholesterol and other sterols control the spatial organization of membrane receptors for influenza and increase viral binding avidity. A natural consequence of this finding is that local cholesterol concentration in the plasma membrane of cells may alter the binding avidity of influenza virions. Furthermore, our results demonstrate a form of cholesterol-dependent membrane organization that does not involve lipid rafts, suggesting that cholesterol's effect on cell membrane heterogeneity is likely the interplay of several different factors.
Conflict of interest statement
Conflicts of interest There are no conflicts of interest to declare.
Figures


Similar articles
-
Influenza viral membrane fusion is sensitive to sterol concentration but surprisingly robust to sterol chemical identity.Sci Rep. 2016 Jul 19;6:29842. doi: 10.1038/srep29842. Sci Rep. 2016. PMID: 27431907 Free PMC article.
-
Target Membrane Cholesterol Modulates Single Influenza Virus Membrane Fusion Efficiency but Not Rate.Biophys J. 2020 May 19;118(10):2426-2433. doi: 10.1016/j.bpj.2020.03.021. Epub 2020 Apr 4. Biophys J. 2020. PMID: 32298636 Free PMC article.
-
A Dynamic, Supramolecular View on the Multivalent Interaction between Influenza Virus and Host Cell.Small. 2021 Apr;17(13):e2007214. doi: 10.1002/smll.202007214. Epub 2021 Mar 7. Small. 2021. PMID: 33682339 Review.
-
Do sterols reduce proton and sodium leaks through lipid bilayers?Prog Lipid Res. 2001 Jul;40(4):299-324. doi: 10.1016/s0163-7827(01)00009-1. Prog Lipid Res. 2001. PMID: 11412894 Review.
-
Hierarchical Multivalent Effects Control Influenza Host Specificity.ACS Cent Sci. 2020 Dec 23;6(12):2311-2318. doi: 10.1021/acscentsci.0c01175. Epub 2020 Nov 12. ACS Cent Sci. 2020. PMID: 33376792 Free PMC article.
Cited by
-
Viral Size Modulates Sendai Virus Binding to Cholesterol-Stabilized Receptor Nanoclusters.J Phys Chem B. 2022 Sep 15;126(36):6802-6810. doi: 10.1021/acs.jpcb.2c03830. Epub 2022 Aug 24. J Phys Chem B. 2022. PMID: 36001793 Free PMC article.
-
Tailored Multivalent Targeting of Siglecs with Photosensitizing Liposome Nanocarriers.Angew Chem Int Ed Engl. 2022 Aug 1;61(31):e202206900. doi: 10.1002/anie.202206900. Epub 2022 Jun 21. Angew Chem Int Ed Engl. 2022. PMID: 35652453 Free PMC article.
-
Molecular Mechanisms behind Conformational Transitions of the Influenza Virus Hemagglutinin Membrane Anchor.J Phys Chem B. 2023 Nov 9;127(44):9450-9460. doi: 10.1021/acs.jpcb.3c05257. Epub 2023 Oct 25. J Phys Chem B. 2023. PMID: 37877534 Free PMC article.
-
Single-Virus Fusion Measurements Reveal Multiple Mechanistically Equivalent Pathways for SARS-CoV-2 Entry.J Virol. 2023 May 31;97(5):e0199222. doi: 10.1128/jvi.01992-22. Epub 2023 May 3. J Virol. 2023. PMID: 37133381 Free PMC article.
-
Apolipoprotein E mediates cell resistance to influenza virus infection.Sci Adv. 2022 Sep 23;8(38):eabm6668. doi: 10.1126/sciadv.abm6668. Epub 2022 Sep 21. Sci Adv. 2022. PMID: 36129973 Free PMC article.
References
-
- Skehel J. J., Wiley D. C. Annu. Rev. Biochem. 2000;69:531–569. - PubMed
-
- Paulson J. C., Sadler J. E., Hill R. L. J. Biol. Chem. 1979;254:2120–2124. - PubMed
-
- Shinya K., Ebina M., Yamada S., Ono M., Kasai N., Kawaoka Y. Nature. 2006;440:435–436. - PubMed
-
- van Riel D., Munster V. J., de Wit E., Rimmelzwaan G. F., Fouchier R. A., Osterhaus A. D., Kuiken T. Science. 2006;312:399. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

