Novel method for DNA methylation analysis using high-performance liquid chromatography and its clinical application
- PMID: 29520901
- PMCID: PMC5980336
- DOI: 10.1111/cas.13566
Novel method for DNA methylation analysis using high-performance liquid chromatography and its clinical application
Abstract
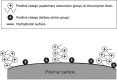
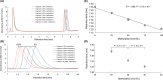
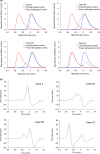
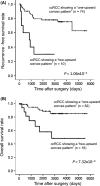
The aim of this study was to develop a new methodology that is suitable for DNA methylation diagnostics and to demonstrate its clinical applicability. We developed a new anion-exchange column for high-performance liquid chromatography (HPLC) with electrostatic and hydrophobic properties. Both cytosine and thymine, corresponding to methylated and unmethylated cytosine after bisulfite modification, respectively, are captured by electrostatic interaction and then discriminated from each other by their hydrophobic interactions. The DNA methylation levels of synthetic DNA were quantified accurately and reproducibly within 10 minutes without time-consuming pretreatment of PCR products, and the measured values were unaffected by the distribution of methylated CpG within the synthetic DNA fragments. When the DNA methylation status of the FAM150A gene, a marker of the CpG island methylator phenotype specific to clear cell renal cell carcinoma (ccRCC), was examined in 98 patients with ccRCC, bulk specimens of tumorous tissue including cancer cells showing DNA methylation of the FAM150A gene were easily identifiable by simply viewing the differentiated chromatograms, even when the cancer cell content was low. Sixteen ccRCC showing DNA methylation more frequently exhibited clinicopathological parameters reflecting tumor aggressiveness (ie, a larger diameter, higher histological grade, vascular involvement, renal vein tumor thrombi, infiltrating growth, tumor necrosis, renal pelvis invasion and higher pathological TNM stage), and had significantly lower recurrence-free and overall survival rates. These data indicate that HPLC analysis using this newly developed anion-exchange column could be a powerful tool for DNA methylation diagnostics, including prognostication of patients with cancers, in a clinical setting.
Keywords: CpG island methylator phenotype; DNA methylation diagnostics; anion-exchange column for high-performance liquid chromatography; clear cell renal cell carcinoma; prognostication.
© 2018 The Authors. Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Figures
Similar articles
-
Prognostication of patients with clear cell renal cell carcinomas based on quantification of DNA methylation levels of CpG island methylator phenotype marker genes.BMC Cancer. 2014 Oct 20;14:772. doi: 10.1186/1471-2407-14-772. BMC Cancer. 2014. PMID: 25332168 Free PMC article.
-
DNA methylation status defines clinicopathological parameters including survival for patients with clear cell renal cell carcinoma (ccRCC).Tumour Biol. 2016 Aug;37(8):10219-28. doi: 10.1007/s13277-016-4893-5. Epub 2016 Jan 30. Tumour Biol. 2016. PMID: 26831665
-
Prognostic significance of Gremlin1 (GREM1) promoter CpG island hypermethylation in clear cell renal cell carcinoma.Am J Pathol. 2010 Feb;176(2):575-84. doi: 10.2353/ajpath.2010.090442. Epub 2009 Dec 30. Am J Pathol. 2010. PMID: 20042676 Free PMC article.
-
Molecular pathological approach to cancer epigenomics and its clinical application.Pathol Int. 2024 Apr;74(4):167-186. doi: 10.1111/pin.13418. Epub 2024 Mar 14. Pathol Int. 2024. PMID: 38482965 Free PMC article. Review.
-
Development of a prognostic risk model for clear cell renal cell carcinoma by systematic evaluation of DNA methylation markers.Clin Epigenetics. 2021 May 4;13(1):103. doi: 10.1186/s13148-021-01084-8. Clin Epigenetics. 2021. PMID: 33947447 Free PMC article.
Cited by
-
DNA Methylation and Non-Coding RNAs during Tissue-Injury Associated Pain.Int J Mol Sci. 2022 Jan 11;23(2):752. doi: 10.3390/ijms23020752. Int J Mol Sci. 2022. PMID: 35054943 Free PMC article. Review.
-
DNA methylation-based classification and identification of renal cell carcinoma prognosis-subgroups.Cancer Cell Int. 2019 Jul 16;19:185. doi: 10.1186/s12935-019-0900-4. eCollection 2019. Cancer Cell Int. 2019. PMID: 31346320 Free PMC article.
-
Prognostication of early-onset endometrioid endometrial cancer based on genome-wide DNA methylation profiles.J Gynecol Oncol. 2022 Nov;33(6):e74. doi: 10.3802/jgo.2022.33.e74. Epub 2022 Aug 16. J Gynecol Oncol. 2022. PMID: 36047377 Free PMC article.
-
Advancements in DNA methylation technologies and their application in cancer diagnosis.Epigenetics. 2025 Dec;20(1):2539995. doi: 10.1080/15592294.2025.2539995. Epub 2025 Jul 28. Epigenetics. 2025. PMID: 40726108 Free PMC article. Review.
-
Quantification of DNA methylation for carcinogenic risk estimation in patients with non-alcoholic steatohepatitis.Clin Epigenetics. 2022 Dec 5;14(1):168. doi: 10.1186/s13148-022-01379-4. Clin Epigenetics. 2022. PMID: 36471401 Free PMC article.
References
-
- Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17:630‐641. - PubMed
-
- Arai E, Kanai Y. DNA methylation profiles in precancerous tissue and cancers: carcinogenetic risk estimation and prognostication based on DNA methylation status. Epigenomics. 2010;2:467‐481. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

