Identification and characterisation of a Theileria annulata proline-rich microtubule and SH3 domain-interacting protein (TaMISHIP) that forms a complex with CLASP1, EB1, and CD2AP at the schizont surface
- PMID: 29520916
- PMCID: PMC6033098
- DOI: 10.1111/cmi.12838
Identification and characterisation of a Theileria annulata proline-rich microtubule and SH3 domain-interacting protein (TaMISHIP) that forms a complex with CLASP1, EB1, and CD2AP at the schizont surface
Abstract
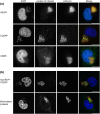
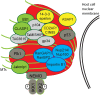
Theileria annulata is an apicomplexan parasite that modifies the phenotype of its host cell completely, inducing uncontrolled proliferation, resistance to apoptosis, and increased invasiveness. The infected cell thus resembles a cancer cell, and changes to various host cell signalling pathways accompany transformation. Most of the molecular mechanisms leading to Theileria-induced immortalization of leukocytes remain unknown. The parasite dissolves the surrounding host cell membrane soon after invasion and starts interacting with host proteins, ensuring its propagation by stably associating with the host cell microtubule network. By using BioID technology together with fluorescence microscopy and co-immunoprecipitation, we identified a CLASP1/CD2AP/EB1-containing protein complex that surrounds the schizont throughout the host cell cycle and integrates bovine adaptor proteins (CIN85, 14-3-3 epsilon, and ASAP1). This complex also includes the schizont membrane protein Ta-p104 together with a novel secreted T. annulata protein (encoded by TA20980), which we term microtubule and SH3 domain-interacting protein (TaMISHIP). TaMISHIP localises to the schizont surface and contains a functional EB1-binding SxIP motif, as well as functional SH3 domain-binding Px(P/A)xPR motifs that mediate its interaction with CD2AP. Upon overexpression in non-infected bovine macrophages, TaMISHIP causes binucleation, potentially indicative of a role in cytokinesis.
Keywords: BioID; CD2AP; Theileria; adaptor proteins; host-parasite interactions; microtubules.
© 2018 The Authors Cellular Microbiology Published by John Wiley & Sons Ltd.
Figures



Similar articles
-
TurboID mapping reveals the exportome of secreted intrinsically disordered proteins in the transforming parasite Theileria annulata.mBio. 2024 Jun 12;15(6):e0341223. doi: 10.1128/mbio.03412-23. Epub 2024 May 15. mBio. 2024. PMID: 38747635 Free PMC article.
-
Recruitment of EB1, a master regulator of microtubule dynamics, to the surface of the Theileria annulata schizont.PLoS Pathog. 2013 May;9(5):e1003346. doi: 10.1371/journal.ppat.1003346. Epub 2013 May 9. PLoS Pathog. 2013. PMID: 23675298 Free PMC article.
-
The Microtubule-Stabilizing Protein CLASP1 Associates with the Theileria annulata Schizont Surface via Its Kinetochore-Binding Domain.mSphere. 2017 Aug 23;2(4):e00215-17. doi: 10.1128/mSphere.00215-17. eCollection 2017 Jul-Aug. mSphere. 2017. PMID: 28861517 Free PMC article.
-
Theileria's Strategies and Effector Mechanisms for Host Cell Transformation: From Invasion to Immortalization.Front Cell Dev Biol. 2021 Apr 20;9:662805. doi: 10.3389/fcell.2021.662805. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33959614 Free PMC article. Review.
-
Alteration of host cell phenotype by Theileria annulata and Theileria parva: mining for manipulators in the parasite genomes.Int J Parasitol. 2006 Jan;36(1):9-21. doi: 10.1016/j.ijpara.2005.09.002. Epub 2005 Sep 30. Int J Parasitol. 2006. PMID: 16221473 Review.
Cited by
-
Screening and identification of Theileria annulata subtelomere-encoded variable secreted protein-950454 (SVSP454) interacting proteins from bovine B cells.Parasit Vectors. 2021 Jun 11;14(1):319. doi: 10.1186/s13071-021-04820-4. Parasit Vectors. 2021. PMID: 34116718 Free PMC article.
-
Dual RNA-seq to catalogue host and parasite gene expression changes associated with virulence of T. annulata-transformed bovine leukocytes: towards identification of attenuation biomarkers.Sci Rep. 2023 Oct 24;13(1):18202. doi: 10.1038/s41598-023-45458-9. Sci Rep. 2023. PMID: 37875584 Free PMC article.
-
Dynamic methylation of histone H3K18 in differentiating Theileria parasites.Nat Commun. 2021 May 28;12(1):3221. doi: 10.1038/s41467-021-23477-2. Nat Commun. 2021. PMID: 34050145 Free PMC article.
-
Recruitment of Host Nuclear Pore Components to the Vicinity of Theileria Schizonts.mSphere. 2020 Feb 5;5(1):e00709-19. doi: 10.1128/mSphere.00709-19. mSphere. 2020. PMID: 32024710 Free PMC article.
-
The Eukaryotic Linear Motif resource: 2022 release.Nucleic Acids Res. 2022 Jan 7;50(D1):D497-D508. doi: 10.1093/nar/gkab975. Nucleic Acids Res. 2022. PMID: 34718738 Free PMC article.
References
-
- Adamson, R. , & Hall, R. (1997). A role for matrix metalloproteinases in the pathology and attenuation of Theileria annulata infections. Parasitol Today (Regul Ed), 13, 390–393. - PubMed
-
- Akhmanova, A. , Hoogenraad, C. C. , Drabek, K. , Stepanova, T. , Dortland, B. , Verkerk, T. , … Galjart, N. (2001). Clasps are CLIP‐115 and ‐170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell, 104, 923–935. - PubMed
-
- Bakheit, M. A. , Ahmed, J. S. , & Seitzer, U. (2008). Existence of splicing variants in homologues of Theileria lestoquardi clone‐5 gene's transcripts in Theileria annulata and Theileria parva. Annals of the New York Academy of Sciences, 1149, 212–213. - PubMed
-
- Bakheit, M. A. , Scholzen, T. , Ahmed, J. S. , & Seitzer, U. (2006). Molecular characterization of a Theileria lestoquardi gene encoding for immunogenic protein splice variants. Parasitology Research, 100, 161–170. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

