Modules of co-occurrence in the cyanobacterial pan-genome reveal functional associations between groups of ortholog genes
- PMID: 29522508
- PMCID: PMC5862535
- DOI: 10.1371/journal.pgen.1007239
Modules of co-occurrence in the cyanobacterial pan-genome reveal functional associations between groups of ortholog genes
Abstract
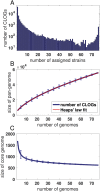
Cyanobacteria are a monophyletic phylogenetic group of global importance and have received considerable attention as potential host organisms for the renewable synthesis of chemical bulk products from atmospheric CO2. The cyanobacterial phylum exhibits enormous metabolic diversity with respect to morphology, lifestyle and habitat. As yet, however, research has mostly focused on few model strains and cyanobacterial diversity is insufficiently understood. In this respect, the increasing availability of fully sequenced bacterial genomes opens new and unprecedented opportunities to investigate the genetic inventory of organisms in the context of their pan-genome. Here, we seek understand cyanobacterial diversity using a comparative genome analysis of 77 fully sequenced and assembled cyanobacterial genomes. We use phylogenetic profiling to analyze the co-occurrence of clusters of likely ortholog genes (CLOGs) and reveal novel functional associations between CLOGs that are not captured by co-localization of genes. Going beyond pair-wise co-occurrences, we propose a network approach that allows us to identify modules of co-occurring CLOGs. The extracted modules exhibit a high degree of functional coherence and reveal known as well as previously unknown functional associations. We argue that the high functional coherence observed for the modules is a consequence of the similar-yet-diverse nature of cyanobacteria. Our approach highlights the importance of a multi-strain analysis to understand gene functions and environmental adaptations, with implications beyond the cyanobacterial phylum. The analysis is augmented with a simple toolbox that facilitates further analysis to investigate the co-occurrence neighborhood of specific CLOGs of interest.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Ducat DC, Way JC, Silver PA. Engineering cyanobacteria to generate high-value products. Trends Biotechnol. 2011;29(2):95–103. doi: 10.1016/j.tibtech.2010.12.003 - DOI - PubMed
-
- Calteau A, Fewer DP, Latifi A, Coursin T, Laurent T, Jokela J, et al. Phylum-wide comparative genomics unravel the diversity of secondary metabolism in Cyanobacteria. BMC Genomics. 2014;15:977 doi: 10.1186/1471-2164-15-977 - DOI - PMC - PubMed
-
- Savakis P, Hellingwerf KJ. Engineering cyanobacteria for direct biofuel production from CO2. Curr Opin Biotechnol. 2015;33:8–14. doi: 10.1016/j.copbio.2014.09.007 - DOI - PubMed
-
- Shih PM, Wu D, Latifi A, Axen SD, Fewer DP, Talla E, et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc Natl Acad Sci U S A. 2013;110(3):1053–8. doi: 10.1073/pnas.1217107110 - DOI - PMC - PubMed
-
- Fujisawa T, Narikawa R, Maeda SI, Watanabe S, Kanesaki Y, Kobayashi K, et al. CyanoBase: a large-scale update on its 20th anniversary. Nucleic Acids Res. 2017;45(D1):D551–D554. doi: 10.1093/nar/gkw1131 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

