Paradoxical roles of ATF6α and ATF6β in modulating disease severity caused by mutations in collagen X
- PMID: 29522813
- PMCID: PMC6090092
- DOI: 10.1016/j.matbio.2018.03.004
Paradoxical roles of ATF6α and ATF6β in modulating disease severity caused by mutations in collagen X
Abstract
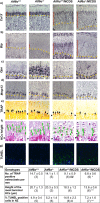
Whilst the role of ATF6α in modulating the unfolded protein response (UPR) has been well documented, the function of its paralogue ATF6β is less well understood. Using knockdown in cell culture and gene ablation in mice we have directly compared the roles of ATF6α & β in responding to the increased ER stress induced by mutant forms of type X collagen that cause the ER stress-associated metaphyseal chondrodysplasia type Schmid (MCDS). ATF6α more efficiently deals with the disease-associated ER stress in the absence of ATF6β and conversely, ATF6β is less effective in the absence of ATF6α. Furthermore, disease severity in vivo is increased by ATF6α ablation and decreased by ATF6β ablation. In addition, novel functions for each paralogue are described including an ATF6β-specific role in controlling growth plate chondrocyte proliferation. The clear demonstration of the intimate relationship of the two ATF6 isoforms and how ATF6β can moderate the activity of ATF6α and vice versa is of great significance for understanding the UPR mechanism. The activities of both ATF6 isoforms and their separate roles need consideration when deciding how to target increased ER stress as a means of treating MCDS and other ER stress-associated diseases.
Copyright © 2018 University of Manchester. Published by Elsevier B.V. All rights reserved.
Figures














Similar articles
-
Partial limitation of cellular functions and compensatory modulation of unfolded protein response pathways caused by double-knockout of ATF6α and ATF6β.Cell Stress Chaperones. 2024 Feb;29(1):34-48. doi: 10.1016/j.cstres.2023.11.002. Epub 2023 Nov 20. Cell Stress Chaperones. 2024. PMID: 38320450 Free PMC article.
-
Carbamazepine reduces disease severity in a mouse model of metaphyseal chondrodysplasia type Schmid caused by a premature stop codon (Y632X) in the Col10a1 gene.Hum Mol Genet. 2018 Nov 15;27(22):3840-3853. doi: 10.1093/hmg/ddy253. Hum Mol Genet. 2018. PMID: 30010889 Free PMC article.
-
Hypertrophic chondrocytes have a limited capacity to cope with increases in endoplasmic reticulum stress without triggering the unfolded protein response.J Histochem Cytochem. 2012 Oct;60(10):734-48. doi: 10.1369/0022155412458436. Epub 2012 Aug 1. J Histochem Cytochem. 2012. PMID: 22859705 Free PMC article.
-
Sledgehammer to Scalpel: Broad Challenges to the Heart and Other Tissues Yield Specific Cellular Responses via Transcriptional Regulation of the ER-Stress Master Regulator ATF6α.Int J Mol Sci. 2020 Feb 8;21(3):1134. doi: 10.3390/ijms21031134. Int J Mol Sci. 2020. PMID: 32046286 Free PMC article. Review.
-
Endoplasmic reticulum stress in chondrodysplasias caused by mutations in collagen types II and X.Cell Stress Chaperones. 2016 Nov;21(6):943-958. doi: 10.1007/s12192-016-0719-z. Epub 2016 Aug 15. Cell Stress Chaperones. 2016. PMID: 27523816 Free PMC article. Review.
Cited by
-
XBP1 signalling is essential for alleviating mutant protein aggregation in ER-stress related skeletal disease.PLoS Genet. 2019 Jul 1;15(7):e1008215. doi: 10.1371/journal.pgen.1008215. eCollection 2019 Jul. PLoS Genet. 2019. PMID: 31260448 Free PMC article.
-
Identification of two novel COL10A1 heterozygous mutations in two Chinese pedigrees with Schmid-type metaphyseal chondrodysplasia.BMC Med Genet. 2019 Dec 19;20(1):200. doi: 10.1186/s12881-019-0937-1. BMC Med Genet. 2019. PMID: 31856751 Free PMC article.
-
Cartilage endoplasmic reticulum stress may influence the onset but not the progression of experimental osteoarthritis.Arthritis Res Ther. 2019 Sep 11;21(1):206. doi: 10.1186/s13075-019-1988-6. Arthritis Res Ther. 2019. PMID: 31511053 Free PMC article.
-
ATF6α contributes to rheumatoid arthritis by inducing inflammatory cytokine production and apoptosis resistance.Front Immunol. 2022 Oct 10;13:965708. doi: 10.3389/fimmu.2022.965708. eCollection 2022. Front Immunol. 2022. PMID: 36300114 Free PMC article.
-
Transcription factors activated through RIP (regulated intramembrane proteolysis) and RAT (regulated alternative translocation).J Biol Chem. 2020 Jul 24;295(30):10271-10280. doi: 10.1074/jbc.REV120.012669. Epub 2020 Jun 2. J Biol Chem. 2020. PMID: 32487748 Free PMC article. Review.
References
-
- Parmar V.M., Schroder M. Sensing endoplasmic reticulum stress. Adv. Exp. Med. Biol. 2012;738:153–168. - PubMed
-
- Dufey E., Sepulveda D., Rojas-Rivera D., Hetz C. Cellular mechanisms of endoplasmic reticulum stress signaling in health and disease. 1. An overview. Am. J. Phys. Cell Phys. 2014 Oct 1;307(7):C582–94. - PubMed
-
- Patterson S.E., Dealy C.N. Mechanisms and models of endoplasmic reticulum stress in chondrodysplasia. Dev. Dyn. 2014 Jul;243(7):875–893. - PubMed
-
- Bateman J.F., Wilson R., Freddi S., Lamande S.R., Savarirayan R. Mutations of COL10A1 in Schmid metaphyseal chondrodysplasia. Hum. Mutat. 2005 Jun;25(6):525–534. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

