Metabolic engineering of the 2-ketobutyrate biosynthetic pathway for 1-propanol production in Saccharomyces cerevisiae
- PMID: 29523149
- PMCID: PMC5844117
- DOI: 10.1186/s12934-018-0883-1
Metabolic engineering of the 2-ketobutyrate biosynthetic pathway for 1-propanol production in Saccharomyces cerevisiae
Abstract
Background: To produce 1-propanol as a potential biofuel, metabolic engineering of microorganisms, such as E. coli, has been studied. However, 1-propanol production using metabolically engineered Saccharomyces cerevisiae, which has an amazing ability to produce ethanol and is thus alcohol-tolerant, has infrequently been reported. Therefore, in this study, we aimed to engineer S. cerevisiae strains capable of producing 1-propanol at high levels.
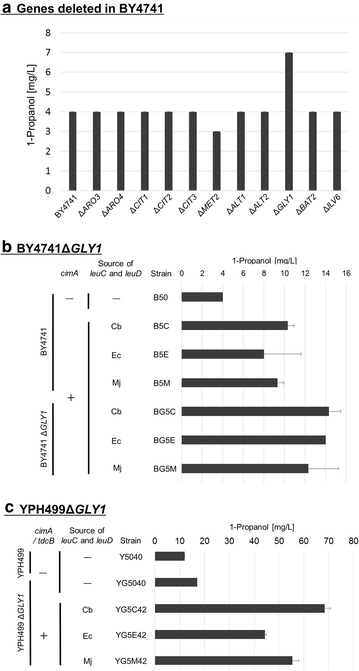
Results: We found that the activity of endogenous 2-keto acid decarboxylase and alcohol/aldehyde dehydrogenase is sufficient to convert 2-ketobutyrate (2 KB) to 500 mg/L 1-propanol in yeast. Production of 1-propanol could be increased by: (i) the construction of an artificial 2 KB biosynthetic pathway from pyruvate via citramalate (cimA); (ii) overexpression of threonine dehydratase (tdcB); (iii) enhancement of threonine biosynthesis from aspartate (thrA, thrB and thrC); and (iv) deletion of the GLY1 gene that regulates a competing pathway converting threonine to glycine. With high-density anaerobic fermentation of the engineered S. cerevisiae strain YG5C4231, we succeeded in producing 180 mg/L 1-propanol from glucose.
Conclusion: These results indicate that the engineering of a citramalate-mediated pathway as a production method for 1-propanol in S. cerevisiae is effective. Although optimization of the carbon flux in S. cerevisiae is necessary to harness this pathway, it is a promising candidate for the large-scale production of 1-propanol.
Keywords: 1-Propanol; 2-Ketobutyrate; Fermentation; S. cerevisiae; Yeast.
Figures






Similar articles
-
Identification of Core Regulatory Genes and Metabolic Pathways for the n-Propanol Synthesis in Saccharomyces cerevisiae.J Agric Food Chem. 2021 Feb 10;69(5):1637-1646. doi: 10.1021/acs.jafc.0c06810. Epub 2021 Jan 27. J Agric Food Chem. 2021. PMID: 33502852
-
Genetic engineering to alter carbon flux for various higher alcohol productions by Saccharomyces cerevisiae for Chinese Baijiu fermentation.Appl Microbiol Biotechnol. 2018 Feb;102(4):1783-1795. doi: 10.1007/s00253-017-8715-5. Epub 2018 Jan 5. Appl Microbiol Biotechnol. 2018. PMID: 29305698
-
Overexpressing enzymes of the Ehrlich pathway and deleting genes of the competing pathway in Saccharomyces cerevisiae for increasing 2-phenylethanol production from glucose.J Biosci Bioeng. 2016 Jul;122(1):34-9. doi: 10.1016/j.jbiosc.2015.12.022. Epub 2016 Mar 11. J Biosci Bioeng. 2016. PMID: 26975754
-
[Metabolic engineering strategies for carboxylic acids production by Saccharomyces cerevisiae---a review].Wei Sheng Wu Xue Bao. 2011 Dec;51(12):1571-7. Wei Sheng Wu Xue Bao. 2011. PMID: 22379797 Review. Chinese.
-
Production of fuels and chemicals from xylose by engineered Saccharomyces cerevisiae: a review and perspective.Microb Cell Fact. 2017 May 11;16(1):82. doi: 10.1186/s12934-017-0694-9. Microb Cell Fact. 2017. PMID: 28494761 Free PMC article. Review.
Cited by
-
Advances in biosynthesis of higher alcohols in Escherichia coli.World J Microbiol Biotechnol. 2023 Mar 21;39(5):125. doi: 10.1007/s11274-023-03580-w. World J Microbiol Biotechnol. 2023. PMID: 36941474 Review.
-
High-Level Production of Isoleucine and Fusel Alcohol by Expression of the Feedback Inhibition-Insensitive Threonine Deaminase in Saccharomyces cerevisiae.Appl Environ Microbiol. 2022 Mar 8;88(5):e0213021. doi: 10.1128/AEM.02130-21. Epub 2022 Jan 12. Appl Environ Microbiol. 2022. PMID: 35020456 Free PMC article.
-
Model-Guided Rational Construction of Escherichia coli Synthetic Consortia for Enhanced 2-Methylbutyric Acid Production.Adv Sci (Weinh). 2025 Jun;12(22):e2416272. doi: 10.1002/advs.202416272. Epub 2025 May 5. Adv Sci (Weinh). 2025. PMID: 40323223 Free PMC article.
-
Production of (R)-citramalate by engineered Saccharomyces cerevisiae.Metab Eng Commun. 2024 Aug 10;19:e00247. doi: 10.1016/j.mec.2024.e00247. eCollection 2024 Dec. Metab Eng Commun. 2024. PMID: 39246525 Free PMC article.
-
Metabolomics-Driven Identification of the Rate-Limiting Steps in 1-Propanol Production.Front Microbiol. 2022 Apr 14;13:871624. doi: 10.3389/fmicb.2022.871624. eCollection 2022. Front Microbiol. 2022. PMID: 35495658 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

