A Decade of Streptococcus thermophilus Phage Evolution in an Irish Dairy Plant
- PMID: 29523549
- PMCID: PMC5930364
- DOI: 10.1128/AEM.02855-17
A Decade of Streptococcus thermophilus Phage Evolution in an Irish Dairy Plant
Abstract
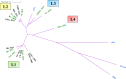
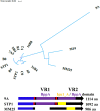
Phages of Streptococcus thermophilus present a major threat to the production of many fermented dairy products. To date, only a few studies have assessed the biodiversity of S. thermophilus phages in dairy fermentations. In order to develop strategies to limit phage predation in this important industrial environment, it is imperative that such studies are undertaken and that phage-host interactions of this species are better defined. The present study investigated the biodiversity and evolution of phages within an Irish dairy fermentation facility over an 11-year period. This resulted in the isolation of 17 genetically distinct phages, all of which belong to the so-called cos group. The evolution of phages within the factory appears to be influenced by phages from other dairy plants introduced into the factory for whey protein powder production. Modular exchange, primarily within the regions encoding lysogeny and replication functions, was the major observation among the phages isolated between 2006 and 2016. Furthermore, the genotype of the first isolate in 2006 was observed continuously across the following decade, highlighting the ability of these phages to prevail in the factory setting for extended periods of time. The proteins responsible for host recognition were analyzed, and carbohydrate-binding domains (CBDs) were identified in the distal tail (Dit), the baseplate proteins, and the Tail-associated lysin (Tal) variable regions (VR1 and VR2) of many isolates. This supports the notion that S. thermophilus phages recognize a carbohydrate receptor on the cell surface of their host.IMPORTANCE Dairy fermentations are consistently threatened by the presence of bacterial viruses (bacteriophages or phages), which may lead to a reduction in acidification rates or even complete loss of the fermentate. These phages may persist in factories for long periods of time. The objective of the current study was to monitor the progression of phages infecting the dairy bacterium Streptococcus thermophilus over a period of 11 years in an Irish dairy plant so as to understand how these phages evolve. A focused analysis of the genomic region that encodes host recognition functions highlighted that the associated proteins harbor a variety of carbohydrate-binding domains, which corroborates the notion that phages of S. thermophilus recognize carbohydrate receptors at the initial stages of the phage cycle.
Keywords: Streptococcus; bacteriophage; dairy industry; genomics; receptor binding protein.
Copyright © 2018 American Society for Microbiology.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

