Quorum-sensing regulator RhlR but not its autoinducer RhlI enables Pseudomonas to evade opsonization
- PMID: 29523648
- PMCID: PMC5934776
- DOI: 10.15252/embr.201744880
Quorum-sensing regulator RhlR but not its autoinducer RhlI enables Pseudomonas to evade opsonization
Abstract
When Drosophila melanogaster feeds on Pseudomonas aeruginosa, some bacteria cross the intestinal barrier and eventually proliferate in the hemocoel. This process is limited by hemocytes through phagocytosis. P. aeruginosa requires the quorum-sensing regulator RhlR to elude the cellular immune response of the fly. RhlI synthesizes the autoinducer signal that activates RhlR. Here, we show that rhlI mutants are unexpectedly more virulent than rhlR mutants, both in fly and in nematode intestinal infection models, suggesting that RhlR has RhlI-independent functions. We also report that RhlR protects P. aeruginosa from opsonization mediated by the Drosophila thioester-containing protein 4 (Tep4). RhlR mutant bacteria show higher levels of Tep4-mediated opsonization, as compared to rhlI mutants, which prevents lethal bacteremia in the Drosophila hemocoel. In contrast, in a septic model of infection, in which bacteria are introduced directly into the hemocoel, Tep4 mutant flies are more resistant to wild-type P. aeruginosa, but not to the rhlR mutant. Thus, depending on the infection route, the Tep4 opsonin can either be protective or detrimental to host defense.
Keywords: competition opsonization‐detection by pattern recognition receptors; infection route; intestinal infection; phagocytosis; quorum sensing.
© 2018 The Authors.
Figures

- A
Representative survival curves of infected and non‐infected (NI) flies. Flies died faster from the infection with PA14 WT than with rhlR. Flies infected with rhlI exhibited an intermediate survival phenotype. One representative experiment is shown. Statistical analysis of the data is shown in (B).
- B
Pooled LT50 data of wild‐type flies (w A5001) following intestinal infections with PA14 WT, rhlR, or rhlI. LT50 of flies after infection with PA14 WT was significantly lower than with rhlR or rhlI (***P < 0.0001 and *P = 0.0009, respectively). Flies were significantly more susceptible to infection with rhlI than with rhlR. The LT50 data from seven survival experiments are displayed (biological duplicates are also shown as there was as much variability between experiments as within experiments). ***P < 0.001.
- C, D
Bacterial titer of the hemolymph collected from flies that had ingested wild‐type or mutant PA14 as indicated, three (C) or five (D) days after the ingestion of PA14 WT or ingestion of mutants as indicated. In this series of experiments, flies infected with PA14 WT had started to succumb by day 5 and were therefore not analyzed. ***P < 0.001.
- E
Survival curves of wild‐type and latex bead‐injected flies after intestinal infection with PA14 bacteria. In latex bead‐injected flies, rhlI regained virulence. Note, however, that the shift in virulence was of the same amplitude as that observed for PA14 WT and contrasts with the large shift observed with rhlR.
- F
Pooled LT50 data of latex bead‐injected flies (w‐LXB) survival experiments. w‐LxB flies died significantly slower after rhlR infection than with PA14 WT (*P = 0.015). A slight decrease in virulence was observed between PA14 WT and rhlI (*P = 0.02). No difference in virulence was detected between rhlR and rhlI. Data represent the LT50s from five experiments (biological duplicates are also shown as there was as much variability between experiments as within experiments).
- G
Differences of LT50s between WT flies pre‐injected with PBS (WT, filled circles; these data are also shown in B) and flies pre‐injected with latex beads (LXB, open circles; these data are also shown in F) after intestinal infection with PA14 WT, rhlR, or rhlI, bars indicate medians. Δ represents the difference between PBS and LXB‐injected flies (LT50[wt‐wtLXB]), and this difference was highly significant for all bacterial genotypes (P = 0.0032 for WT PA14 and rhlI, P < 0.0001 for rhlR; not shown on the graph for simplicity). Data represent the LT50s from five experiments (biological duplicates are also shown as there was as much variability between experiments as within experiments). *P < 0.05.
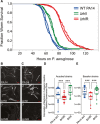
- A
An automated C. elegans lifespan machine was used to monitor the survival of worms in a P. aeruginosa‐mediated killing “slow‐killing” assay 64. Approximately 200 wild‐type C. elegans nematodes were fed WT PA14 (blue), rhlI (ΔrhlI: green), or rhlR (ΔrhlR: red) (constructed in the Ausubel or Bassler laboratories, hence two curves for each genotype); P < 0.001 (log‐rank test) for PA14 versus rhlI, PA14 versus rhlR, or rhlI versus rhlR. The experiment was repeated twice with similar results.
- B–E
C. elegans wild‐type N2 animals were fed WT PA14, rhlI, or rhlR (constructed in the Ausubel (B and D) or Bassler (C and E) laboratories expressing GFP). At 48 h post‐infection, 13–18 worms infected with WT PA14, rhlI, or rhlR were imaged in the green fluorescent channel. (B, C) Representative images are shown. Scale bar is 200 μm. (D, E) Images were quantified using ImageJ. There was a significant difference in the levels of live bacteria between WT PA14 and rhlR (***P = 0.0003, D and E), and there were significant differences between the rhlR and rhlI mutants (***P = 8 × 10−7 (D) and 0.0005 (E)). The data fitted a negative binomial distribution and were analyzed using generalized linear models by one‐way ANOVA. The differences between rhlI and WT PA14 were not significant. The experiments were repeated at least two times with similar results. Bars represent medians, the upper and lower limits of boxes indicate, respectively, the first and third quartiles and whiskers encompass data points within 1.5 times the interquartile range.

- A, B
Wild‐type Drosophila were orally infected with wild‐type PA14 expressing dsRed (PA14‐dsRed). After 4 days, infected flies were transferred to tubes containing wild‐type PA14 expressing GFP (PA14‐GFP). At day 5 of the infection (1 day after transferring flies to PA14‐GFP), most PA14 bacteria in the Drosophila gut expressed dsRed and only 10% expressed GFP (A). However, at day 6 (2 days after the transfer of flies to PA14‐GFP), only GFP‐positive bacteria were detected in the gut. Results were similar for bacteria retrieved from the hemolymph compartment (B). Each dot corresponds to a sample of 10 flies.
- C–E
LT50s from survival experiments of WT flies (w) after intestinal infection with PA14 WT (C), rhlR (D), or rhlI (E) mutants and injection of either latex beads (LXB, open circles) or PBS (filled circles) at different time points of the infection. Latex beads or PBS was injected either 1 day before the infection started (−1 day) or 4 h (+4 h), 1 day (+1 day), 4 days (+4 days), or 6 days (+6 days) after the infection started. Black filled circles correspond to the survival of infected, uninjected flies. (C) LT50s of w A5001‐LxB were significantly lower than w A5001 only at −1 day (**P = 0.003) and +4 h (*P = 0.013). (D) LT50s of w‐LXB flies were significantly lower than w at most times during the infection (−1 day: ***P = 5 × 10−5, +4 h: ***P = 3 × 10−6, and +4 day: **P = 0.01). (E) A similar phenotype was observed with flies infected with rhlI (−1 day: **P = 0.002, +4 h: **P = 0.006, +1 day: ***P = 8 × 10−6, and +4 days: *P = 0.03). Note, however, that for injections of latex beads at day 4, the difference was reduced, as compared to earlier time points of injection of latex beads. The cumulative LT50 data from at least three experiments (only two experiments for rhlI) are shown, except for day 6.
- F
Δ: Differences of LT50s between WT flies injected with PBS (WT) and flies injected with latex beads (WTLXB) after intestinal infection with PA14 WT, rhlR, or rhlI in at least two experiments. *P < 0.05; ***P < 0.001.
- G
Guts of transgenic flies with GFP‐labeled hemocytes were dissected in a manner that preserves the association of hemocytes with the digestive tract and were examined by fluorescence confocal microscopy. Green: GFP; blue: DAPI staining of nuclei. Scale bars: 100 μm.
- H
Analysis of hemocytes recruited to the fly intestine upon infection with either wild‐type PA14, rhlR, or rhlI. All three strains elicited recruitment of hemocytes to the gut (4 h after the beginning of the infection, for each bacteria ***P < 0.0001); at 3 days after the beginning of the infection, there were fewer hemocytes recruited after infection with rhlI (**P = 0.0025 between rhlR and rhlI). Data represent three pooled experiments. *P < 0.05.

- A, B
Drosophila wild‐type flies (w A5001), single Tep3 or Tep4 mutants, the double Tep2,3 mutant, and the triple Tep2,3,4 mutant were orally infected with PA14 WT (A) or rhlR (B) in parallel survival experiments. (A) Tep4 and Tep2,3,4 mutant flies were significantly more susceptible to PA14 infection compared to w A5001. No difference in survival was detected between the Tep2,3 mutant and w A5001. Surprisingly, Tep3 mutants seemed to be more resistant to infection. (B) A strong enhancement of rhlR virulence was observed with the Tep4 and Tep2,3,4 mutants compared to w A5001 flies. Tep2,3 and w A5001 exhibited nearly the same rate of death when challenged with rhlR. The Tep3 mutant seemed again to be more resistant to the infection. One representative experiment out of three (each with biological triplicates, except for uninfected flies) is shown.
- C
The survival of Tep4 flies infected with PA14 WT, rhlR, or rhlI was examined. One representative experiment out of three (each with biological triplicates) is shown.
- D
Quantification of the experiments shown in (C), which had been performed in parallel with w A5001 flies; filled circles: wild‐type flies, open circles: Tep4. The triplicates were analyzed as independent experiments as there was as much variability between experiments as within experiments. All differences between wild‐type flies and the Tep4 mutant (Δ) were highly significant (***P < 0.0001). Bars represent medians, the upper and lower limits of boxes indicate, respectively, the first and third quartiles and whiskers encompass data points within 1.5 times the interquartile range. Data were analyzed using linear models and reported P‐values are relative to the post‐hoc pairwise comparisons between the groups of interest. **P < 0.01; ***P < 0.001.

Heat‐killed pHrodo®‐labeled bacteria of the indicated genotype were injected into either wild‐type or Tep4 third‐instar larvae and incubated for 45 min. The hemocytes were then retrieved. Bacteria present in phagosomes were fluorescent and used to measure the phagocytic index. rhlR bacteria injected into Tep4 (open circles) were significantly more phagocytosed than wild‐type bacteria (filled circles) (P = 0.02); PA14 WT were more readily phagocytosed by wild type than by Tep4 hemocytes (*P = 0.01).
The experiment is similar to that shown in (A), except that live bacteria were used and a differential antibody staining procedure was performed to reveal phagocytosed bacteria: Phagocytosed bacteria were stained only after permeabilization and fluoresced green whereas non‐ingested bacteria were stained by both secondary antibodies, red, and green, which were, respectively, used prior and after the permeabilization step (see Materials and Methods section). PA14 WT, rhlR, or rhlI were injected in wild‐type or Tep4 larvae. Again, PA14 WT was more readily phagocytosed by wild‐type than by Tep4 larval hemocytes (***P = 0.0006). rhlR were engulfed more efficiently than PA14 WT bacteria by Tep4 hemocytes (*P = 0.01).

- A
Scheme of the experimental procedure. Live bacteria were incubated with either wild‐type or Tep4 hemolymph and were thereafter injected into naive larvae, which were either wild‐type or Tep4. The phagocytosis index was then measured as in Fig 4B.
- B, C
Bacteria pre‐incubated with wild‐type or Tep4 hemolymph are represented in pairs, respectively, with filled (left) and open (right) circles. PA14 WT, rhlR, or rhlI bacteria pre‐incubated with either wild‐type (wt) or Tep4 hemolymph were injected into wild‐type larvae (B) or into Tep4 larvae (C). Data were analyzed using generalized linear models assuming a negative binomial distribution and statistically homogeneous groups (indicated by letters) were defined based on the post‐hoc pairwise comparisons between the different groups. The difference between each group is highly significant (P < 0.001). Bars represent medians, the upper and lower limits of boxes indicate, respectively, the first and third quartiles and whiskers encompass data points within 1.5 times the interquartile range.

- A, B
PA14 WT, rhlR, or a 1:1 mix thereof was submitted to the opsonization assay using wild‐type larvae as donors and recipients. Panel (A) displays the measured phagocytic indexes: PA14 WT is hardly ingested by larval hemocytes, whereas rhlR and the mixture of wild‐type and rhlR bacteria display a similar distribution, as shown in (B), where the cumulative distribution of phagocytic indices is shown. The purple curve (PA14_WT_ΔrhlR_join) shows the distribution that would be obtained by pooling the data from rhlR and PA14 WT alone in this opsonization assay. The observed distribution of the phagocytic indices in the mix is clearly closer to that of rhlR (two‐sample Kolmogorov‐Smirnov test, P = 0.98), suggesting that the ingested bacteria are mostly of the rhlR genotype. Bars represent medians, the upper and lower limits of boxes indicate, respectively, the first and third quartiles and whiskers encompass data points within 1.5 times the interquartile range.

- A, B
The survival of wild‐type and Tep4 flies was examined, after injection of PBS as a non‐infected control (NI), or 10, 100, 1,000 CFUs of PA14 WT (A) or rhlR (B). One representative experiment out of 10 (A) or out of three (B) is shown. **P < 0.01; ***P < 0.001. A log‐rank test was used.
- C
Diptericin expression was measured by RT–qPCR in wild‐type and Tep4 flies at 2, 8, or 24 h after injection of PBS (NI), or of 10, 100, 1,000 CFUs of PA14 WT bacteria. Each experiment was performed independently three times, and a representative experiment is shown. The Diptericin expression shown is relative to RpL32 expression and normalized to the non‐infected wild‐type control. Error bars represent the standard deviation. *P < 0.05; **P < 0.01 (unpaired t‐test).
- D
The cleavage of the prophenoloxidase was analyzed 2 h after injection of 1,000 CFUs of PA14 WT bacteria or PBS, by Western blotting using a pan‐phenoloxidase antibody. The intensity of the prophenoloxidase (PPO) and of the cleaved active phenoloxidase (activated PO) bands was measured, and the ratio of the measurements is shown below the blot. This experiment was performed twice with a similar result.
Similar articles
-
Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family.J Bacteriol. 1995 Dec;177(24):7155-63. doi: 10.1128/jb.177.24.7155-7163.1995. J Bacteriol. 1995. PMID: 8522523 Free PMC article.
-
Pseudomonas aeruginosa RhlR is required to neutralize the cellular immune response in a Drosophila melanogaster oral infection model.Proc Natl Acad Sci U S A. 2011 Oct 18;108(42):17378-83. doi: 10.1073/pnas.1114907108. Epub 2011 Oct 10. Proc Natl Acad Sci U S A. 2011. PMID: 21987808 Free PMC article.
-
A rhlI 5' UTR-Derived sRNA Regulates RhlR-Dependent Quorum Sensing in Pseudomonas aeruginosa.mBio. 2019 Oct 8;10(5):e02253-19. doi: 10.1128/mBio.02253-19. mBio. 2019. PMID: 31594819 Free PMC article.
-
Pseudomonas aeruginosa quorum sensing modulates immune responses: An updated review article.Immunol Lett. 2017 Oct;190:1-6. doi: 10.1016/j.imlet.2017.07.002. Epub 2017 Jul 8. Immunol Lett. 2017. PMID: 28698104 Review.
-
An evolving perspective on the Pseudomonas aeruginosa orphan quorum sensing regulator QscR.Front Cell Infect Microbiol. 2014 Oct 28;4:152. doi: 10.3389/fcimb.2014.00152. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 25389523 Free PMC article. Review.
Cited by
-
Nora virus proliferates in dividing intestinal stem cells and sensitizes flies to intestinal infection and oxidative stress.bioRxiv [Preprint]. 2025 Feb 4:2025.01.30.635658. doi: 10.1101/2025.01.30.635658. bioRxiv. 2025. PMID: 39975242 Free PMC article. Preprint.
-
Haemocyte-mediated immunity in insects: Cells, processes and associated components in the fight against pathogens and parasites.Immunology. 2021 Nov;164(3):401-432. doi: 10.1111/imm.13390. Epub 2021 Aug 2. Immunology. 2021. PMID: 34233014 Free PMC article. Review.
-
Production of quorum sensing-related metabolites and phytoalexins during Pseudomonas aeruginosa-Brassica napus interaction.Microbiology (Reading). 2022 Aug;168(8):001212. doi: 10.1099/mic.0.001212. Microbiology (Reading). 2022. PMID: 35980361 Free PMC article.
-
A specific innate immune response silences the virulence of Pseudomonas aeruginosa in a latent infection model in the Drosophila melanogaster host.PLoS Pathog. 2024 Jun 4;20(6):e1012252. doi: 10.1371/journal.ppat.1012252. eCollection 2024 Jun. PLoS Pathog. 2024. PMID: 38833496 Free PMC article.
-
An updated proteomic analysis of Drosophila haemolymph after bacterial infection.Fly (Austin). 2025 Dec;19(1):2485685. doi: 10.1080/19336934.2025.2485685. Epub 2025 Apr 13. Fly (Austin). 2025. PMID: 40223358 Free PMC article.
References
-
- Grimont PA, Grimont F (1978) The genus Serratia. Annu Rev Microbiol 32: 221–248 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

