Structure-function analyses of a stereotypic rheumatoid factor unravel the structural basis for germline-encoded antibody autoreactivity
- PMID: 29523691
- PMCID: PMC5936827
- DOI: 10.1074/jbc.M117.814475
Structure-function analyses of a stereotypic rheumatoid factor unravel the structural basis for germline-encoded antibody autoreactivity
Abstract
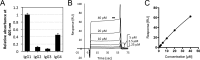
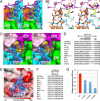
Rheumatoid factors (RFs) are autoantibodies against the fragment-crystallizable (Fc) region of IgG. In individuals with hematological diseases such as cryoglobulinemia and certain B cell lymphoma forms, the RFs derived from specific heavy- and light-chain germline pairs, so-called "stereotypic RFs," are frequently produced in copious amounts and form immune complexes with IgG in serum. Of note, many structural details of the antigen recognition mechanisms in RFs are unclear. Here we report the crystal structure of the RF YES8c derived from the IGHV1-69/IGKV3-20 germline pair, the most common of the stereotypic RFs, in complex with human IgG1-Fc at 2.8 Å resolution. We observed that YES8c binds to the CH2-CH3 elbow in the canonical antigen-binding manner involving a large antigen-antibody interface. On the basis of this observation, combined with mutational analyses, we propose a recognition mechanism common to IGHV1-69/IGKV3-20 RFs: (1) the interaction of the Leu432-His435 region of Fc enables the highly variable complementarity-determining region (CDR)-H3 to participate in the binding, (2) the hydrophobic tip in the CDR-H2 typical of IGHV1-69 antibodies recognizes the hydrophobic patch on Fc, and (3) the interaction of the highly conserved RF light chain with Fc is important for RF activity. These features may determine the putative epitope common to the IGHV1-69/IGKV3-20 RFs. We also showed that some mutations in the binding site of RF increase the affinity to Fc, which may aggravate hematological diseases. Our findings unravel the structural basis for germline-encoded antibody autoreactivity.
Keywords: IgG; autoimmunity; complex; crystal structure; structural biology.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



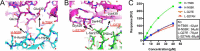
Similar articles
-
Structural Basis of a Conventional Recognition Mode of IGHV1-69 Rheumatoid Factors.Adv Exp Med Biol. 2021;21:171-182. doi: 10.1007/5584_2020_510. Adv Exp Med Biol. 2021. PMID: 32185699 Review.
-
The structure of a human rheumatoid factor bound to IgG Fc.Adv Exp Med Biol. 1998;435:41-50. doi: 10.1007/978-1-4615-5383-0_4. Adv Exp Med Biol. 1998. PMID: 9498063 Review.
-
Crystal structure of a human autoimmune complex between IgM rheumatoid factor RF61 and IgG1 Fc reveals a novel epitope and evidence for affinity maturation.J Mol Biol. 2007 May 18;368(5):1321-31. doi: 10.1016/j.jmb.2007.02.085. Epub 2007 Mar 6. J Mol Biol. 2007. PMID: 17395205 Free PMC article.
-
Isolation, characterization, and molecular modeling of a rheumatoid factor from a Hepatitis C virus infected patient with Sjögren's syndrome.ScientificWorldJournal. 2013 Dec 30;2013:516516. doi: 10.1155/2013/516516. eCollection 2013. ScientificWorldJournal. 2013. PMID: 24489505 Free PMC article.
-
Salivary Gland Mucosa-Associated Lymphoid Tissue-Type Lymphoma From Sjögren's Syndrome Patients in the Majority Express Rheumatoid Factors Affinity-Selected for IgG.Arthritis Rheumatol. 2020 Aug;72(8):1330-1340. doi: 10.1002/art.41263. Epub 2020 Jul 8. Arthritis Rheumatol. 2020. PMID: 32182401 Free PMC article.
Cited by
-
COVID-19, varying genetic resistance to viral disease and immune tolerance checkpoints.Immunol Cell Biol. 2021 Feb;99(2):177-191. doi: 10.1111/imcb.12419. Epub 2020 Nov 23. Immunol Cell Biol. 2021. PMID: 33113212 Free PMC article. Review.
-
Autoantibodies - enemies, and/or potential allies?Front Immunol. 2022 Oct 19;13:953726. doi: 10.3389/fimmu.2022.953726. eCollection 2022. Front Immunol. 2022. PMID: 36341384 Free PMC article. Review.
-
Production of scFv, Fab, and IgG of CR3022 Antibodies Against SARS-CoV-2 Using Silkworm-Baculovirus Expression System.Mol Biotechnol. 2021 Dec;63(12):1223-1234. doi: 10.1007/s12033-021-00373-0. Epub 2021 Jul 25. Mol Biotechnol. 2021. PMID: 34304364 Free PMC article.
-
Peripheral B cells from patients with hepatitis C virus-associated lymphoma exhibit clonal expansion and an anergic-like transcriptional profile.iScience. 2022 Dec 15;26(1):105801. doi: 10.1016/j.isci.2022.105801. eCollection 2023 Jan 20. iScience. 2022. PMID: 36619973 Free PMC article.
-
Testing for isotypes does not help differentiating rheumatoid arthritis from other rheumatoid factor positive diseases.Immunol Res. 2023 Dec;71(6):883-886. doi: 10.1007/s12026-023-09402-3. Epub 2023 Jun 15. Immunol Res. 2023. PMID: 37322352
References
-
- Randen I., Thompson K. M., Pascual V., Victor K., Beale D., Coadwell J., Førre O., Capra J. D., and Natvig J. B. (1992) Rheumatoid factor V genes from patients with rheumatoid arthritis are diverse and show evidence of an antigen-driven response. Immunol. Rev. 128, 49–71 10.1111/j.1600-065X.1992.tb00832.x - DOI - PubMed
-
- Pascual V., Randen I., Thompson K., Sioud M., Forre O., Natvig J., and Capra J. D. (1990) The complete nucleotide sequences of the heavy chain variable regions of six monospecific rheumatoid factors derived from Epstein-Barr virus-transformed B cells isolated from the synovial tissue of patients with rheumatoid arthritis: further evidence that some autoantibodies are unmutated copies of germ line genes. J. Clin. Invest. 86, 1320–1328 10.1172/JCI114841 - DOI - PMC - PubMed
-
- Victor K. D., Randen I., Thompson K., Forre O., Natvig J. B., Fu S. M., and Capra J. D. (1991) Rheumatoid factors isolated from patients with autoimmune disorders are derived from germline genes distinct from those encoding the Wa, Po, and Bla cross-reactive idiotypes. J. Clin. Invest. 87, 1603–1613 10.1172/JCI115174 - DOI - PMC - PubMed
-
- Pascual V., Victor K., Randen I., Thompson K., Steinitz M., Førre O., Fu S. M., Natvig J. B., and Capra J. D. (1992) Nucleotide sequence analysis of rheumatoid factors and polyreactive antibodies derived from patients with rheumatoid arthritis reveals diverse use of VH and VL gene segments and extensive variability in CDR-3. Scand. J. Immunol. 36, 349–362 10.1111/j.1365-3083.1992.tb03108.x - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

