FERTILIZATION-INDEPENDENT SEED-Polycomb Repressive Complex 2 Plays a Dual Role in Regulating Type I MADS-Box Genes in Early Endosperm Development
- PMID: 29523711
- PMCID: PMC5933120
- DOI: 10.1104/pp.17.00534
FERTILIZATION-INDEPENDENT SEED-Polycomb Repressive Complex 2 Plays a Dual Role in Regulating Type I MADS-Box Genes in Early Endosperm Development
Abstract
Early endosperm development presents a unique system in which to uncover epigenetic regulatory mechanisms because the contributing maternal and paternal genomes possess differential epigenetic modifications. In Arabidopsis (Arabidopsis thaliana), the initiation of endosperm coenocytic growth upon fertilization and the transition to endosperm cellularization are regulated by the FERTILIZATION-INDEPENDENT SEED (FIS)-Polycomb Repressive Complex 2 (PRC2), a putative H3K27 methyltransferase. Here, we address the possible role of the FIS-PRC2 complex in regulating the type I MADS-box gene family, which has been shown previously to regulate early endosperm development. We show that a subclass of type I MADS-box genes (C2 genes) was expressed in distinct domains of the coenocytic endosperm in wild-type seeds. Furthermore, the C2 genes were mostly up-regulated biallelically during the extended coenocytic phase of endosperm development in the FIS-PRC2 mutant background. Using allele-specific expression analysis, we also identified a small subset of C2 genes subjected to FIS-PRC2-dependent maternal or FIS-PRC2-independent paternal imprinting. Our data support a dual role for the FIS-PRC2 complex in the regulation of C2 type I MADS-box genes, as evidenced by a generalized role in the repression of gene expression at both alleles associated with endosperm cellularization and a specialized role in silencing the maternal allele of imprinted genes.
© 2018 American Society of Plant Biologists. All Rights Reserved.
Figures



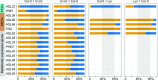
 and
and  (where m denotes maternal transcripts and p denotes paternal transcripts measured by SBE) in both reciprocal crosses, as described previously (Hsieh et al., 2011). P values calculated using Student’s t test are provided in
(where m denotes maternal transcripts and p denotes paternal transcripts measured by SBE) in both reciprocal crosses, as described previously (Hsieh et al., 2011). P values calculated using Student’s t test are provided in 
Similar articles
-
Two MADS-box proteins, AGL9 and AGL15, recruit the FIS-PRC2 complex to trigger the phase transition from endosperm proliferation to embryo development in Arabidopsis.Mol Plant. 2024 Jul 1;17(7):1110-1128. doi: 10.1016/j.molp.2024.05.011. Epub 2024 Jun 1. Mol Plant. 2024. PMID: 38825830
-
Genome-wide transcript profiling of endosperm without paternal contribution identifies parent-of-origin-dependent regulation of AGAMOUS-LIKE36.PLoS Genet. 2011 Feb;7(2):e1001303. doi: 10.1371/journal.pgen.1001303. Epub 2011 Feb 17. PLoS Genet. 2011. PMID: 21379330 Free PMC article.
-
The Polycomb group protein MEDEA and the DNA methyltransferase MET1 interact to repress autonomous endosperm development in Arabidopsis.Plant J. 2013 Mar;73(5):776-87. doi: 10.1111/tpj.12070. Epub 2013 Feb 12. Plant J. 2013. PMID: 23146178
-
Epigenetic mechanisms governing seed development in plants.EMBO Rep. 2006 Dec;7(12):1223-7. doi: 10.1038/sj.embor.7400854. EMBO Rep. 2006. PMID: 17139298 Free PMC article. Review.
-
[Imprinting genes and it's expression in Arabidopsis].Yi Chuan. 2010 Jul;32(7):670-6. doi: 10.3724/sp.j.1005.2010.00670. Yi Chuan. 2010. PMID: 20650847 Review. Chinese.
Cited by
-
The co-expression of genes involved in seed coat and endosperm development promotes seed abortion in grapevine.Planta. 2021 Sep 28;254(5):87. doi: 10.1007/s00425-021-03728-8. Planta. 2021. PMID: 34585280
-
Characterization analyses of MADS-box genes highlighting their functions with seed development in Ricinus communis.Front Plant Sci. 2025 May 14;16:1589915. doi: 10.3389/fpls.2025.1589915. eCollection 2025. Front Plant Sci. 2025. PMID: 40438739 Free PMC article.
-
Family plot: the impact of the endosperm and other extra-embryonic seed tissues on angiosperm zygotic embryogenesis.F1000Res. 2020 Jan 14;9:F1000 Faculty Rev-18. doi: 10.12688/f1000research.21527.1. eCollection 2020. F1000Res. 2020. PMID: 32055398 Free PMC article. Review.
-
The phenomenon of autonomous endosperm in sexual and apomictic plants.J Exp Bot. 2023 Aug 17;74(15):4324-4348. doi: 10.1093/jxb/erad168. J Exp Bot. 2023. PMID: 37155961 Free PMC article. Review.
-
The maternally expressed polycomb group gene OsEMF2a is essential for endosperm cellularization and imprinting in rice.Plant Commun. 2020 Jul 2;2(1):100092. doi: 10.1016/j.xplc.2020.100092. eCollection 2021 Jan 11. Plant Commun. 2020. PMID: 33511344 Free PMC article.
References
-
- Adams S, Vinkenoog R, Spielman M, Dickinson HG, Scott RJ (2000) Parent-of-origin effects on seed development in Arabidopsis thaliana require DNA methylation. Development 127: 2493–2502 - PubMed
-
- Baud S, Wuillème S, Lemoine R, Kronenberger J, Caboche M, Lepiniec L, Rochat C (2005) The AtSUC5 sucrose transporter specifically expressed in the endosperm is involved in early seed development in Arabidopsis. Plant J 43: 824–836 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

