Exploiting CELLULOSE SYNTHASE (CESA) Class Specificity to Probe Cellulose Microfibril Biosynthesis
- PMID: 29523715
- PMCID: PMC5933121
- DOI: 10.1104/pp.18.00263
Exploiting CELLULOSE SYNTHASE (CESA) Class Specificity to Probe Cellulose Microfibril Biosynthesis
Abstract
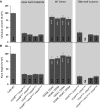
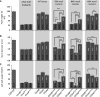
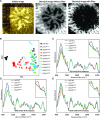
Cellulose microfibrils are the basic units of cellulose in plants. The structure of these microfibrils is at least partly determined by the structure of the cellulose synthase complex. In higher plants, this complex is composed of 18 to 24 catalytic subunits known as CELLULOSE SYNTHASE A (CESA) proteins. Three different classes of CESA proteins are required for cellulose synthesis and for secondary cell wall cellulose biosynthesis these classes are represented by CESA4, CESA7, and CESA8. To probe the relationship between CESA proteins and microfibril structure, we created mutant cesa proteins that lack catalytic activity but retain sufficient structural integrity to allow assembly of the cellulose synthase complex. Using a series of Arabidopsis (Arabidopsis thaliana) mutants and genetic backgrounds, we found consistent differences in the ability of these mutant cesa proteins to complement the cellulose-deficient phenotype of the cesa null mutants. The best complementation was observed with catalytically inactive cesa4, while the equivalent mutation in cesa8 exhibited significantly lower levels of complementation. Using a variety of biophysical techniques, including solid-state nuclear magnetic resonance and Fourier transform infrared microscopy, to study these mutant plants, we found evidence for changes in cellulose microfibril structure, but these changes largely correlated with cellulose content and reflected differences in the relative proportions of primary and secondary cell walls. Our results suggest that individual CESA classes have similar roles in determining cellulose microfibril structure, and it is likely that the different effects of mutating members of different CESA classes are the consequence of their different catalytic activity and their influence on the overall rate of cellulose synthesis.
© 2018 American Society of Plant Biologists. All Rights Reserved.
Figures






Similar articles
-
The three cellulose synthase isoforms for secondary cell wall make specific contributions to microfibril synthesis.Plant J. 2025 Jul;123(2):e70344. doi: 10.1111/tpj.70344. Plant J. 2025. PMID: 40671415
-
Cellulose microfibril crystallinity is reduced by mutating C-terminal transmembrane region residues CESA1A903V and CESA3T942I of cellulose synthase.Proc Natl Acad Sci U S A. 2012 Mar 13;109(11):4098-103. doi: 10.1073/pnas.1200352109. Epub 2012 Feb 28. Proc Natl Acad Sci U S A. 2012. PMID: 22375033 Free PMC article.
-
Features of the primary wall CESA complex in wild type and cellulose-deficient mutants of Arabidopsis thaliana.J Exp Bot. 2008;59(10):2627-37. doi: 10.1093/jxb/ern125. Epub 2008 May 20. J Exp Bot. 2008. PMID: 18495638 Free PMC article.
-
Cellulose synthases and synthesis in Arabidopsis.Mol Plant. 2011 Mar;4(2):199-211. doi: 10.1093/mp/ssq079. Epub 2011 Feb 9. Mol Plant. 2011. PMID: 21307367 Review.
-
Cellulose microfibrils in plants: biosynthesis, deposition, and integration into the cell wall.Int Rev Cytol. 2000;199:161-99. doi: 10.1016/s0074-7696(00)99004-1. Int Rev Cytol. 2000. PMID: 10874579 Review.
Cited by
-
On the Mechanism of the Ionizing Radiation-Induced Degradation and Recycling of Cellulose.Polymers (Basel). 2023 Nov 22;15(23):4483. doi: 10.3390/polym15234483. Polymers (Basel). 2023. PMID: 38231912 Free PMC article. Review.
-
Cellulose Synthase Stoichiometry in Aspen Differs from Arabidopsis and Norway Spruce.Plant Physiol. 2018 Jul;177(3):1096-1107. doi: 10.1104/pp.18.00394. Epub 2018 May 14. Plant Physiol. 2018. PMID: 29760198 Free PMC article.
-
Five amino acid mismatches in the zinc-finger domains of Cellulose Synthase 5 and Cellulose Synthase 6 cooperatively modulate their functional properties by controlling homodimerization in Arabidopsis.Plant Mol Biol. 2024 Jun 18;114(4):76. doi: 10.1007/s11103-024-01471-8. Plant Mol Biol. 2024. PMID: 38888655
-
Phenotypic effects of changes in the FTVTxK region of an Arabidopsis secondary wall cellulose synthase compared with results from analogous mutations in other isoforms.Plant Direct. 2021 Aug 5;5(8):e335. doi: 10.1002/pld3.335. eCollection 2021 Aug. Plant Direct. 2021. PMID: 34386691 Free PMC article.
-
Genome-wide association and selection studies for pod dehiscence resistance in the USDA soybean germplasm collection.PLoS One. 2025 Mar 28;20(3):e0318815. doi: 10.1371/journal.pone.0318815. eCollection 2025. PLoS One. 2025. PMID: 40153708 Free PMC article.
References
-
- Atalla RH, Vanderhart DL (1984) Native cellulose: a composite of two distinct crystalline forms. Science 223: 283–285 - PubMed
-
- Blakeney AB, Harris PJ, Henry RJ, Stone BA (1983) A simple and rapid preparation of alditol acetates for monosaccharide analysis. Carbohydr Res 113: 291–299
-
- Burton RA, Gidley MJ, Fincher GB (2010) Heterogeneity in the chemistry, structure and function of plant cell walls. Nat Chem Biol 6: 724–732 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

