Protein Acetylation and Butyrylation Regulate the Phenotype and Metabolic Shifts of the Endospore-forming Clostridium acetobutylicum
- PMID: 29523768
- PMCID: PMC5986239
- DOI: 10.1074/mcp.RA117.000372
Protein Acetylation and Butyrylation Regulate the Phenotype and Metabolic Shifts of the Endospore-forming Clostridium acetobutylicum
Abstract
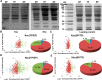
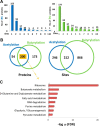
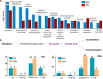
Clostridium acetobutylicum is a strict anaerobic, endospore-forming bacterium, which is used for the production of the high energy biofuel butanol in metabolic engineering. The life cycle of C. acetobutylicum can be divided into two phases, with acetic and butyric acids being produced in the exponential phase (acidogenesis) and butanol formed in the stationary phase (solventogenesis). During the transitional phase from acidogenesis to solventogenesis and latter stationary phase, concentration peaks of the metabolic intermediates butyryl phosphate and acetyl phosphate are observed. As an acyl group donor, acyl-phosphate chemically acylates protein substrates. However, the regulatory mechanism of lysine acetylation and butyrylation involved in the phenotype and solventogenesis of C. acetobutylicum remains unknown. In our study, we conducted quantitative analysis of protein acetylome and butyrylome to explore the dynamic change of lysine acetylation and butyrylation in the exponential phase, transitional phase, and stationary phase of C. acetobutylicum Total 458 lysine acetylation sites and 1078 lysine butyrylation sites were identified in 254 and 373 substrates, respectively. Bioinformatics analysis uncovered the similarities and differences between the two acylation modifications in C. acetobutylicum Mutation analysis of butyrate kinase and the central transcriptional factor Spo0A was performed to characterize the unique role of lysine butyrylation in the metabolic pathway and sporulation process of C. acetobutylicum Moreover, quantitative proteomic assays were performed to reveal the relationship between protein features (e.g. gene expression level and lysine acylation level) and metabolites in the three growth stages. This study expanded our knowledge of lysine acetylation and butyrylation in Clostridia and constituted a resource for functional studies on lysine acylation in bacteria.
Keywords: Acetylation*; Bacteria; Enzyme Regulation*; Mass Spectrometry; Post-translational modifications*; Quantification; acylation regulation; endospore-forming Clostridium; metabolic shift; quantitative acetylome; quantitative butyrylome.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




Similar articles
-
Enhanced butanol production obtained by reinforcing the direct butanol-forming route in Clostridium acetobutylicum.mBio. 2012 Oct 23;3(5):e00314-12. doi: 10.1128/mBio.00314-12. mBio. 2012. PMID: 23093384 Free PMC article.
-
A proteomic and transcriptional view of acidogenic and solventogenic steady-state cells of Clostridium acetobutylicum in a chemostat culture.Appl Microbiol Biotechnol. 2010 Aug;87(6):2209-26. doi: 10.1007/s00253-010-2741-x. Epub 2010 Jul 9. Appl Microbiol Biotechnol. 2010. PMID: 20617312 Free PMC article.
-
Transcriptional program of early sporulation and stationary-phase events in Clostridium acetobutylicum.J Bacteriol. 2005 Oct;187(20):7103-18. doi: 10.1128/JB.187.20.7103-7118.2005. J Bacteriol. 2005. PMID: 16199581 Free PMC article.
-
Recent advances in n-butanol and butyrate production using engineered Clostridium tyrobutyricum.World J Microbiol Biotechnol. 2020 Aug 14;36(9):138. doi: 10.1007/s11274-020-02914-2. World J Microbiol Biotechnol. 2020. PMID: 32794091 Review.
-
Transcriptional regulation of solventogenesis in Clostridium acetobutylicum.J Mol Microbiol Biotechnol. 2002 May;4(3):295-300. J Mol Microbiol Biotechnol. 2002. PMID: 11931561 Review.
Cited by
-
Acetylomics reveals an extensive acetylation diversity within Pseudomonas aeruginosa.Microlife. 2024 Sep 14;5:uqae018. doi: 10.1093/femsml/uqae018. eCollection 2024. Microlife. 2024. PMID: 39464744 Free PMC article.
-
Lysine Phoshoglycerylation Is Widespread in Bacteria and Overlaps with Acylation.Microorganisms. 2024 Jul 30;12(8):1556. doi: 10.3390/microorganisms12081556. Microorganisms. 2024. PMID: 39203397 Free PMC article.
-
Characterization of a novel + 70 Da modification in rhGM-CSF expressed in E. coli using chemical assays in combination with mass spectrometry.Amino Acids. 2022 Apr;54(4):601-613. doi: 10.1007/s00726-021-03004-9. Epub 2021 Aug 28. Amino Acids. 2022. PMID: 34453584 Free PMC article.
-
Recent Contributions of Proteomics to Our Understanding of Reversible Nε-Lysine Acylation in Bacteria.J Proteome Res. 2024 Aug 2;23(8):2733-2749. doi: 10.1021/acs.jproteome.3c00912. Epub 2024 Mar 5. J Proteome Res. 2024. PMID: 38442041 Free PMC article. Review.
-
Mechanisms, Detection, and Relevance of Protein Acetylation in Prokaryotes.mBio. 2019 Apr 9;10(2):e02708-18. doi: 10.1128/mBio.02708-18. mBio. 2019. PMID: 30967470 Free PMC article. Review.
References
-
- Pietrocola F., Galluzzi L., Pedro J. M. B., Madeo F., and Kroemer G. (2015) Acetyl coenzyme A: a central metabolite and second messenger. Cell Metabolism 21, 805–821 - PubMed
-
- Menzies K. J., Zhang H., Katsyuba E., and Auwerx J. (2015) Protein acetylation in metabolism - metabolites and cofactors. Nat. Rev. Endocrinol. 12, 43–60 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

