MRN complex-dependent recruitment of ubiquitylated BLM helicase to DSBs negatively regulates DNA repair pathways
- PMID: 29523790
- PMCID: PMC5844875
- DOI: 10.1038/s41467-018-03393-8
MRN complex-dependent recruitment of ubiquitylated BLM helicase to DSBs negatively regulates DNA repair pathways
Abstract
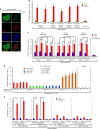
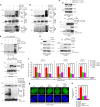
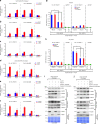
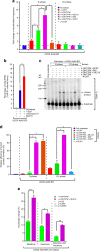
Mutations in BLM in Bloom Syndrome patients predispose them to multiple types of cancers. Here we report that BLM is recruited in a biphasic manner to annotated DSBs. BLM recruitment is dependent on the presence of NBS1, MRE11 and ATM. While ATM activity is essential for BLM recruitment in early phase, it is dispensable in late phase when MRE11 exonuclease activity and RNF8-mediated ubiquitylation of BLM are the key determinants. Interaction between polyubiquitylated BLM and NBS1 is essential for the helicase to be retained at the DSBs. The helicase activity of BLM is required for the recruitment of HR and c-NHEJ factors onto the chromatin in S- and G1-phase, respectively. During the repair phase, BLM inhibits HR in S-phase and c-NHEJ in G1-phase. Consequently, inhibition of helicase activity of BLM enhances the rate of DNA alterations. Thus BLM utilizes its pro- and anti-repair functions to maintain genome stability.
Conflict of interest statement
The authors declare no competing interests.
Figures


Comment in
-
BLM's balancing act and the involvement of FANCJ in DNA repair.Cell Cycle. 2018;17(18):2207-2220. doi: 10.1080/15384101.2018.1520567. Epub 2018 Sep 23. Cell Cycle. 2018. PMID: 30209988 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

