Oat evolution revealed in the maternal lineages of 25 Avena species
- PMID: 29523798
- PMCID: PMC5844911
- DOI: 10.1038/s41598-018-22478-4
Oat evolution revealed in the maternal lineages of 25 Avena species
Abstract
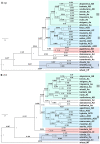
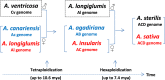
Cultivated hexaploid oat has three different sets of nuclear genomes (A, C, D), but its evolutionary history remains elusive. A multiplexed shotgun sequencing procedure was explored to acquire maternal phylogenetic signals from chloroplast and mitochondria genomes of 25 Avena species. Phylogenetic analyses of the acquired organelle SNP data revealed a new maternal pathway towards hexaploids of oat genome evolution involving three diploid species (A. ventricosa, A. canariensis and A. longiglumis) and two tetraploid species (A. insularis and A. agadiriana). Cultivated hexaploid A. sativa acquired its maternal genome from an AC genome tetraploid close to A. insularis. Both AC genome A. insularis and AB genome A. agadiriana obtained a maternal genome from an ancient A, not C, genome diploid close to A. longiglumis. Divergence dating showed the major divergences of C genome species 19.9-21.2 million years ago (Mya), of the oldest A genome A. canariensis 13-15 Mya, and of the clade with hexaploids 8.5-9.5 Mya. These findings not only advance our knowledge on oat genome evolution, but also have implications for oat germplasm conservation and utilization in breeding.
Conflict of interest statement
The author declares no competing interests.
Figures


Similar articles
-
A study on the genetic relationships of Avena taxa and the origins of hexaploid oat.Theor Appl Genet. 2016 Jul;129(7):1405-1415. doi: 10.1007/s00122-016-2712-4. Epub 2016 Apr 5. Theor Appl Genet. 2016. PMID: 27048238
-
Discrimination of the closely related A and D genomes of the hexaploid oat Avena sativa L.Proc Natl Acad Sci U S A. 1998 Oct 13;95(21):12450-5. doi: 10.1073/pnas.95.21.12450. Proc Natl Acad Sci U S A. 1998. PMID: 9770506 Free PMC article.
-
AFLP variation in 25 Avena species.Theor Appl Genet. 2008 Aug;117(3):333-42. doi: 10.1007/s00122-008-0778-3. Epub 2008 May 7. Theor Appl Genet. 2008. PMID: 18461302
-
[Interspecific crosses in Avena L. species].Genetika. 2001 May;37(5):581-90. Genetika. 2001. PMID: 11436548 Review. Russian.
-
Genotyping-by-Sequencing and Its Application to Oat Genomic Research.Methods Mol Biol. 2017;1536:169-187. doi: 10.1007/978-1-4939-6682-0_13. Methods Mol Biol. 2017. PMID: 28132151 Review.
Cited by
-
Phylogenetic relationships in the genus Avena based on the nuclear Pgk1 gene.PLoS One. 2018 Nov 8;13(11):e0200047. doi: 10.1371/journal.pone.0200047. eCollection 2018. PLoS One. 2018. PMID: 30408035 Free PMC article.
-
New evidence confirming the CD genomic constitutions of the tetraploid Avena species in the section Pachycarpa Baum.PLoS One. 2021 Jan 8;16(1):e0240703. doi: 10.1371/journal.pone.0240703. eCollection 2021. PLoS One. 2021. PMID: 33417607 Free PMC article.
-
Low-coverage whole-genome sequencing facilitates accurate and cost-effective haplotype reconstruction in complex mouse crosses.Mamm Genome. 2025 Jul 1:10.1007/s00335-025-10148-6. doi: 10.1007/s00335-025-10148-6. Online ahead of print. Mamm Genome. 2025. PMID: 40593333 Free PMC article.
-
Comparative chloroplast genome analyses of Avena: insights into evolutionary dynamics and phylogeny.BMC Plant Biol. 2020 Sep 2;20(1):406. doi: 10.1186/s12870-020-02621-y. BMC Plant Biol. 2020. PMID: 32878602 Free PMC article.
-
The comparison of polymorphism among Avena species revealed by retrotransposon-based DNA markers and soluble carbohydrates in seeds.J Appl Genet. 2023 May;64(2):247-264. doi: 10.1007/s13353-023-00748-w. Epub 2023 Jan 31. J Appl Genet. 2023. PMID: 36719514 Free PMC article.
References
-
- Strychar, R. World Oat Production, Trade, and Usage. In: Webster F, Wood P, editors. Oats: Chemistry and Technology. Minnesota, USA: AACC International, Inc. p. 1–10 (2011).
-
- Baum, B.R. Oats: wild and cultivated. A monograph of the genus Avena L. (Poaceae). Minister of Supply and Services, Ottawa (1977).
-
- Ladizinsky G. A new species of Avena from Sicily, possibly the tetraploid progenitor of hexaploid oats. Genet Resour Crop Evol. 1998;45:263–269. doi: 10.1023/A:1008657530466. - DOI
-
- Loskutov IG. On evolutionary pathways of Avena species. Genet Resour Crop Evol. 2008;55:211–220. doi: 10.1007/s10722-007-9229-2. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

