Risk of nontyphoidal Salmonella bacteraemia in African children is modified by STAT4
- PMID: 29523850
- PMCID: PMC5844948
- DOI: 10.1038/s41467-017-02398-z
Risk of nontyphoidal Salmonella bacteraemia in African children is modified by STAT4
Abstract
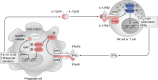
Nontyphoidal Salmonella (NTS) is a major cause of bacteraemia in Africa. The disease typically affects HIV-infected individuals and young children, causing substantial morbidity and mortality. Here we present a genome-wide association study (180 cases, 2677 controls) and replication analysis of NTS bacteraemia in Kenyan and Malawian children. We identify a locus in STAT4, rs13390936, associated with NTS bacteraemia. rs13390936 is a context-specific expression quantitative trait locus for STAT4 RNA expression, and individuals carrying the NTS-risk genotype demonstrate decreased interferon-γ (IFNγ) production in stimulated natural killer cells, and decreased circulating IFNγ concentrations during acute NTS bacteraemia. The NTS-risk allele at rs13390936 is associated with protection against a range of autoimmune diseases. These data implicate interleukin-12-dependent IFNγ-mediated immunity as a determinant of invasive NTS disease in African children, and highlight the shared genetic architecture of infectious and autoimmune disease.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

