A Dominant Negative Antisense Approach Targeting β-Catenin
- PMID: 29524201
- PMCID: PMC5918491
- DOI: 10.1007/s12033-018-0058-7
A Dominant Negative Antisense Approach Targeting β-Catenin
Abstract
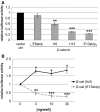
There have been many attempts to unveil the therapeutic potential of antisense molecules during the last decade. Due to its specific role in canonical Wnt signalling, β-catenin is a potential target for an antisense-based antitumour therapy. In order to establish such a strategy with peptide nucleic acids, we developed a reporter assay for quantification of antisense effects. The luciferase-based assay detects splice blocking with high sensitivity. Using this assay, we show that the splice donor of exon 13 of β-catenin is particularly suitable for an antisense strategy, as it results in a truncated protein which lacks transactivating functions. Since the truncated proteins retain the interactions with Tcf/Lef proteins, they act in a dominant negative fashion competing with wild-type proteins and thus blocking the transcriptional activity of β-catenin. Furthermore, we show that the truncation does not interfere with binding of cadherin and α-catenin, both essential for its function in cell adhesion. Therefore, the antisense strategy blocks Wnt signalling with high efficiency but retains other important functions of β-catenin.
Keywords: Antisense; Morpholino; PNA; Wnt signalling; β-Catenin.
Conflict of interest statement
MA, HB, BW and TL were full employees of the company ugichem GmbH that developed PNAs for therapeutic applications. Design and synthesis of the PNAs were performed at the ugichem GmbH, which became insolvent 2015.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

