Identification and Rescue of Splice Defects Caused by Two Neighboring Deep-Intronic ABCA4 Mutations Underlying Stargardt Disease
- PMID: 29526278
- PMCID: PMC5985352
- DOI: 10.1016/j.ajhg.2018.02.008
Identification and Rescue of Splice Defects Caused by Two Neighboring Deep-Intronic ABCA4 Mutations Underlying Stargardt Disease
Abstract
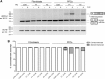
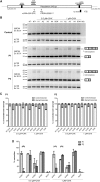
Sequence analysis of the coding regions and splice site sequences in inherited retinal diseases is not able to uncover ∼40% of the causal variants. Whole-genome sequencing can identify most of the non-coding variants, but their interpretation is still very challenging, in particular when the relevant gene is expressed in a tissue-specific manner. Deep-intronic variants in ABCA4 have been associated with autosomal-recessive Stargardt disease (STGD1), but the exact pathogenic mechanism is unknown. By generating photoreceptor precursor cells (PPCs) from fibroblasts obtained from individuals with STGD1, we demonstrated that two neighboring deep-intronic ABCA4 variants (c.4539+2001G>A and c.4539+2028C>T) result in a retina-specific 345-nt pseudoexon insertion (predicted protein change: p.Arg1514Leufs∗36), likely due to the creation of exonic enhancers. Administration of antisense oligonucleotides (AONs) targeting the 345-nt pseudoexon can significantly rescue the splicing defect observed in PPCs of two individuals with these mutations. Intriguingly, an AON that is complementary to c.4539+2001G>A rescued the splicing defect only in PPCs derived from an individual with STGD1 with this but not the other mutation, demonstrating the high specificity of AONs. In addition, a single AON molecule rescued splicing defects associated with different neighboring mutations, thereby providing new strategies for the treatment of persons with STGD1. As many genes associated with human genetic conditions are expressed in specific tissues and pre-mRNA splicing may also rely on organ-specific factors, our approach to investigate and treat splicing variants using differentiated cells derived from individuals with STGD1 can be applied to any tissue of interest.
Keywords: ABCA4; Stargardt disease; antisense oligonucleotide; deep-intronic mutation; exonic splicing enhancer; induced pluripotent stem cells; nonsense-mediated decay; photoreceptor precursor cells; pseudoexon; splicing modulation.
Copyright © 2018 American Society of Human Genetics. All rights reserved.
Figures


References
-
- Fishman G.A., Stone E.M., Grover S., Derlacki D.J., Haines H.L., Hockey R.R. Variation of clinical expression in patients with Stargardt dystrophy and sequence variations in the ABCR gene. Arch. Ophthalmol. 1999;117:504–510. - PubMed
-
- Stargardt K. Über familiäre, progressive Degeneration in der Maculagegend des Auges. Graefes Arch. Clin. Exp. Ophthalmol. 1909;71:534–550.
-
- Allikmets R., Singh N., Sun H., Shroyer N.F., Hutchinson A., Chidambaram A., Gerrard B., Baird L., Stauffer D., Peiffer A. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat. Genet. 1997;15:236–246. - PubMed
-
- Molday R.S., Beharry S., Ahn J., Zhong M. Binding of N-retinylidene-PE to ABCA4 and a model for its transport across membranes. Adv. Exp. Med. Biol. 2006;572:465–470. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

