Gene Correction Reverses Ciliopathy and Photoreceptor Loss in iPSC-Derived Retinal Organoids from Retinitis Pigmentosa Patients
- PMID: 29526738
- PMCID: PMC5998840
- DOI: 10.1016/j.stemcr.2018.02.003
Gene Correction Reverses Ciliopathy and Photoreceptor Loss in iPSC-Derived Retinal Organoids from Retinitis Pigmentosa Patients
Erratum in
-
Gene Correction Reverses Ciliopathy and Photoreceptor Loss in iPSC-Derived Retinal Organoids from Retinitis Pigmentosa Patients.Stem Cell Reports. 2018 Jun 5;10(6):2005. doi: 10.1016/j.stemcr.2018.05.012. Stem Cell Reports. 2018. PMID: 29874627 Free PMC article. No abstract available.
Abstract
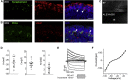
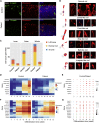
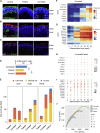
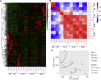
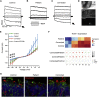
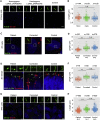
Retinitis pigmentosa (RP) is an irreversible, inherited retinopathy in which early-onset nyctalopia is observed. Despite the genetic heterogeneity of RP, RPGR mutations are the most common causes of this disease. Here, we generated induced pluripotent stem cells (iPSCs) from three RP patients with different frameshift mutations in the RPGR gene, which were then differentiated into retinal pigment epithelium (RPE) cells and well-structured retinal organoids possessing electrophysiological properties. We observed significant defects in photoreceptor in terms of morphology, localization, transcriptional profiling, and electrophysiological activity. Furthermore, shorted cilium was found in patient iPSCs, RPE cells, and three-dimensional retinal organoids. CRISPR-Cas9-mediated correction of RPGR mutation rescued photoreceptor structure and electrophysiological property, reversed the observed ciliopathy, and restored gene expression to a level in accordance with that in the control using transcriptome-based analysis. This study recapitulated the pathogenesis of RPGR using patient-specific organoids and achieved targeted gene therapy of RPGR mutations in a dish as proof-of-concept evidence.
Keywords: RPE cells; RPGR; ciliopathy; cilium; disease modeling; electrophysiology; patient-derived iPSCs; photoreceptor; retinal organoid; retinitis pigmentosa.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


Comment in
-
Gene corrections in sight.Nat Cell Biol. 2018 Jun;20(6):634. doi: 10.1038/s41556-018-0117-0. Nat Cell Biol. 2018. PMID: 29784915 No abstract available.
References
-
- Brunner S., Skosyrski S., Kirschner-Schwabe R., Knobeloch K.P., Neidhardt J., Feil S., Glaus E., Luhmann U.F., Rüther K., Berger W. Cone versus rod disease in a mutant Rpgr mouse caused by different genetic backgrounds. Invest. Ophthalmol. Vis. Sci. 2010;51:1106–1115. - PubMed
-
- Cheong C., Sung Y.H., Lee J., Choi Y.S., Song J., Kee C., Lee H.W. Role of INK4a locus in normal eye development and cataract genesis. Mech. Ageing Dev. 2006;127:633–638. - PubMed
-
- Daiger, S.P., Sullivan, L.S., and Rossiter, B.J.F. (2013). RetNet. The Retinal Information Network. https://sph.uth.edu/retnet/.
-
- Eiraku M., Takata N., Ishibashi H., Kawada M., Sakakura E., Okuda S., Sekiguchi K., Adachi T., Sasai Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472:51–56. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

