Revising the Structural Diversity of Ribosomal Proteins Across the Three Domains of Life
- PMID: 29529322
- PMCID: PMC5995209
- DOI: 10.1093/molbev/msy021
Revising the Structural Diversity of Ribosomal Proteins Across the Three Domains of Life
Abstract
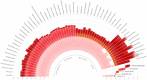
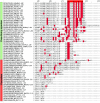
Ribosomal proteins are indispensable components of a living cell, and yet their structures are remarkably diverse in different species. Here we use manually curated structural alignments to provide a comprehensive catalog of structural variations in homologous ribosomal proteins from bacteria, archaea, eukaryotes, and eukaryotic organelles. By resolving numerous ambiguities and errors of automated structural and sequence alignments, we uncover a whole new class of structural variations that reside within seemingly conserved segments of ribosomal proteins. We then illustrate that these variations reflect an apparent adaptation of ribosomal proteins to the specific environments and lifestyles of living species. Finally, we show that most of these structural variations reside within nonglobular extensions of ribosomal proteins-protein segments that are thought to promote ribosome biogenesis by stabilizing the proper folding of ribosomal RNA. We show that although the extensions are thought to be the most ancient peptides on our planet, they are in fact the most rapidly evolving and most structurally and functionally diverse segments of ribosomal proteins. Overall, our work illustrates that, despite being long considered as slowly evolving and highly conserved, ribosomal proteins are more complex and more specialized than is generally recognized.
Figures



References
-
- Agrawal RK, Sharma MR, Yassin A, Lahiri I, Spremulli LL (2011). Structure and function of organellar ribosomes as revealed by cryo-EM In: Rodnina MV, Wintermeyer W, Green R, editors. Ribosomes: structure, function, and dynamics. Springer Link.
-
- Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R.. 2013. Structures of the human and Drosophila 80S ribosome. Nature 4977447:80–85. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

