Prevalence of Plasmodium falciparum delayed clearance associated polymorphisms in adaptor protein complex 2 mu subunit (pfap2mu) and ubiquitin specific protease 1 (pfubp1) genes in Ghanaian isolates
- PMID: 29530100
- PMCID: PMC5848568
- DOI: 10.1186/s13071-018-2762-3
Prevalence of Plasmodium falciparum delayed clearance associated polymorphisms in adaptor protein complex 2 mu subunit (pfap2mu) and ubiquitin specific protease 1 (pfubp1) genes in Ghanaian isolates
Abstract
Background: Plasmodium falciparum delayed clearance with the use of artemisinin-based combination therapy (ACTs) has been reported in some African countries. Single nucleotide polymorphisms (SNPs) in two genes, P. falciparum adaptor protein complex 2 mu subunit (pfap2mu) and ubiquitin specific protease 1 (pfubp1), have been linked to delayed clearance with ACT use in Kenya and recurrent imported malaria in Britain. With over 12 years of ACT use in Ghana, this study investigated the prevalence of SNPs in the pfap2mu and pfubp1 in Ghanaian clinical P. falciparum isolates to provide baseline data for antimalarial drug resistance surveillance in the country.
Methods: Filter paper blood blots collected in 2015-2016 from children aged below 9 years presenting with uncomplicated malaria at hospitals in three sentinel sites Begoro, Cape Coast and Navrongo were used. Parasite DNA was extracted from 120 samples followed by nested polymerase chain reaction (nPCR). Sanger sequencing was performed to detect and identify SNPs in pfap2mu and pfubp1 genes.
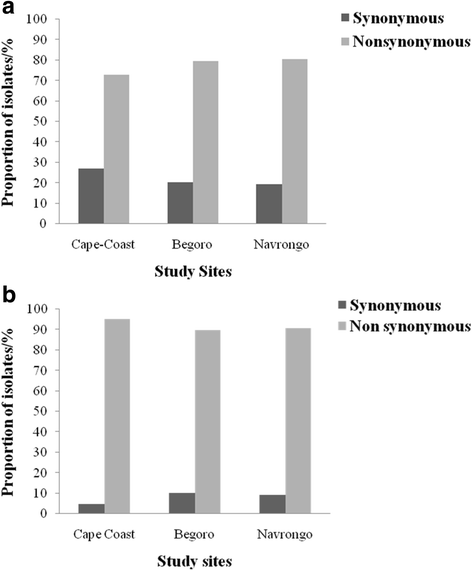
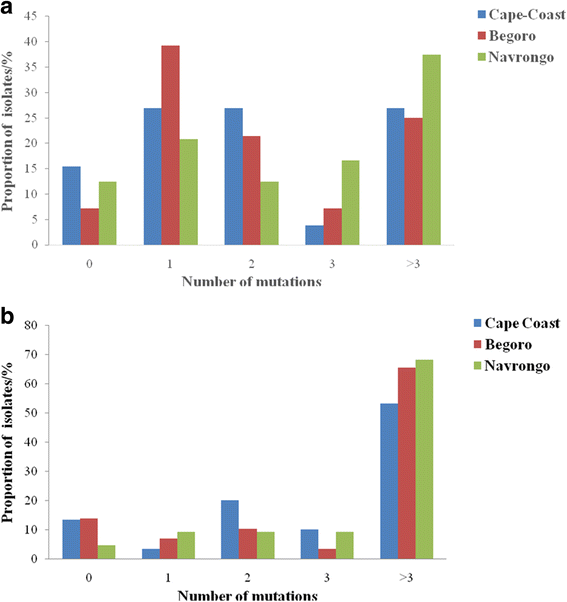
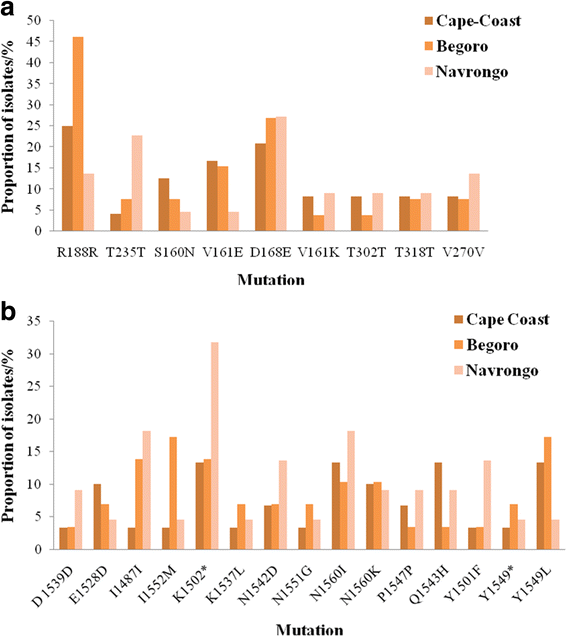
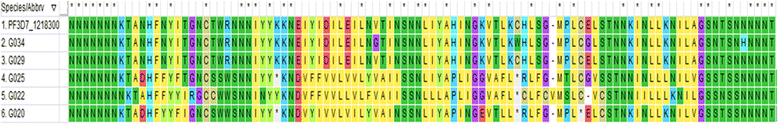
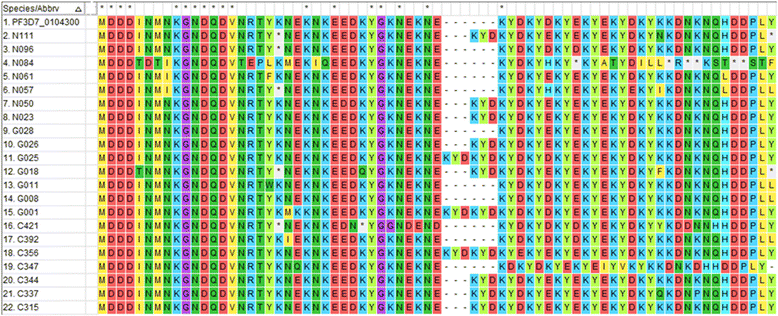
Results: In all, 11.1% (9/81) of the isolates carried the wildtype genotypes for both genes. A total of 164 pfap2mu mutations were detected in 67 isolates whilst 271 pfubp1 mutations were observed in 72 isolates. The majority of the mutations were non-synonymous (NS): 78% (128/164) for pfap2mu and 92.3% (250/271) for pfubp1. Five unique samples had a total of 215 pfap2mu SNPs, ranging between 15 and 63 SNPs per sample. Genotypes reportedly associated with ART resistance detected in this study included pfap2mu S160N (7.4%, 6/81) and pfubp1 E1528D (7.4%, 6/81) as well as D1525E (4.9%, 4/81). There was no significant difference in the prevalence of the SNPs between the three ecologically distinct study sites (pfap2mu: χ2 = 6.905, df = 2, P = 0.546; pfubp1: χ2 = 4.883, df = 2, P = 0.769).
Conclusions: The detection of pfap2mu and pfubp1 genotypes associated with ACT delayed parasite clearance is evidence of gradual nascent emergence of resistance in Ghana. The results will serve as baseline data for surveillance and the selection of the genotypes with drug pressure over time. The pfap2mu S160N, pfubp1 E1528D and D1525E must be monitored in Ghanaian isolates in ACT susceptibility studies, especially when cure rates of ACTs, particularly AL, is less than 100%.
Keywords: ACT; Antimalarial drug resistance; Artemisinin; Ghana; Mutations; Plasmodium falciparum; pfap2mu; pfubp1.
Conflict of interest statement
Ethics approval and consent to participate
Ethical approval for the study was given by the NMIMR Institutional Review Board (IRB). The samples were taken after parents or guardians of the children gave their consent. This work is part of an ongoing surveillance of antimalarial drug efficacy studies in Ghana approved by the NMIMR IRB (CPN032/05-06a).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
-
- WHO. Malaria Fact Sheet 2017. http://www.who.int/mediacentre/factsheets/fs094/en/. Accessed 08 June 2017.
-
- WHO. Malaria Fact Sheet: World Malaria Day 2016. www.who.int/malaria/media/world-malaria-day-2016/en. Accessed 22 Nov 2016.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

