Repair of DNA double-strand breaks by mammalian alternative end-joining pathways
- PMID: 29530982
- PMCID: PMC6036210
- DOI: 10.1074/jbc.TM117.000375
Repair of DNA double-strand breaks by mammalian alternative end-joining pathways
Abstract
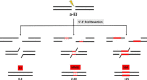
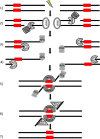
Alternative end-joining (a-EJ) pathways, which repair DNA double-strand breaks (DSBs), are initiated by end resection that generates 3' single strands. This reaction is shared, at least in part, with homologous recombination but distinguishes a-EJ from the major nonhomologous end-joining pathway. Although the a-EJ pathways make only a minor and poorly understood contribution to DSB repair in nonmalignant cells, there is growing interest in these pathways, as they generate genomic rearrangements that are hallmarks of cancer cells. Here, we review and discuss the current understanding of the mechanisms and regulation of a-EJ pathways, the role of a-EJ in human disease, and the potential utility of a-EJ as a therapeutic target in cancer.
Keywords: DNA damage; DNA endonuclease; DNA ligation; DNA polymerase; alternative end-joining pathway; chromosomes; deletions; double-strand break; genomic instability; microhomology; single-strand annealing; translocation.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
A. E. T. is a co-inventor on patents that cover the use of DNA ligase inhibitors as anti-cancer agents, and altered expression of DSB repair proteins as biomarkers of increased dependence upon MMEJ
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

