The transpeptidase PbpA and noncanonical transglycosylase RodA of Mycobacterium tuberculosis play important roles in regulating bacterial cell lengths
- PMID: 29530985
- PMCID: PMC5925798
- DOI: 10.1074/jbc.M117.811190
The transpeptidase PbpA and noncanonical transglycosylase RodA of Mycobacterium tuberculosis play important roles in regulating bacterial cell lengths
Abstract
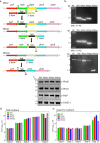
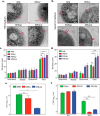
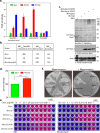
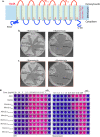
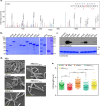
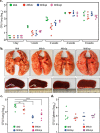
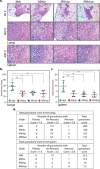
The cell wall of Mycobacterium tuberculosis (Mtb) is a complex structure that protects the pathogen in hostile environments. Peptidoglycan (PG), which helps determine the morphology of the cell envelope, undergoes substantial remodeling under stress. This meshwork of linear chains of sugars, cross-linked through attached peptides, is generated through the sequential action of enzymes termed transglycosylases and transpeptidases. The Mtb genome encodes two classical transglycosylases and four transpeptidases, the functions of which are not fully elucidated. Here, we present work on the yet uncharacterized transpeptidase PbpA and a nonclassical transglycosylase RodA. We elucidate their roles in regulating in vitro growth and in vivo survival of pathogenic mycobacteria. We find that RodA and PbpA are required for regulating cell length, but do not affect mycobacterial growth. Biochemical analyses show PbpA to be a classical transpeptidase, whereas RodA is identified to be a member of an emerging class of noncanonical transglycosylases. Phosphorylation of RodA at Thr-463 modulates its biological function. In a guinea pig infection model, RodA and PbpA are found to be required for both bacterial survival and formation of granuloma structures, thus underscoring the importance of these proteins in mediating mycobacterial virulence in the host. Our results emphasize the fact that whereas redundant enzymes probably compensate for the absence of RodA or PbpA during in vitro growth, the two proteins play critical roles for the survival of the pathogen inside its host.
Keywords: Lipid II; Mycobacterium tuberculosis; PbpA; RodA; bacterial pathogenesis; cell division; cell wall; peptidoglycan; transglycosylase; transpeptidase.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

