Direct observation of ultrafast large-scale dynamics of an enzyme under turnover conditions
- PMID: 29531052
- PMCID: PMC5879700
- DOI: 10.1073/pnas.1720448115
Direct observation of ultrafast large-scale dynamics of an enzyme under turnover conditions
Abstract
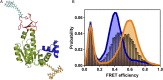
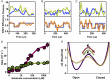
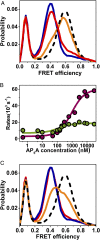
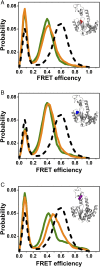
The functional cycle of many proteins involves large-scale motions of domains and subunits. The relation between conformational dynamics and the chemical steps of enzymes remains under debate. Here we show that in the presence of substrates, domain motions of an enzyme can take place on the microsecond time scale, yet exert influence on the much-slower chemical step. We study the domain closure reaction of the enzyme adenylate kinase from Escherichia coli while in action (i.e., under turnover conditions), using single-molecule FRET spectroscopy. We find that substrate binding increases dramatically domain closing and opening times, making them as short as ∼15 and ∼45 µs, respectively. These large-scale conformational dynamics are likely the fastest measured to date, and are ∼100-200 times faster than the enzymatic turnover rate. Some active-site mutants are shown to fully or partially prevent the substrate-induced increase in domain closure times, while at the same time they also reduce enzymatic activity, establishing a clear connection between the two phenomena, despite their disparate time scales. Based on these surprising observations, we propose a paradigm for the mode of action of enzymes, in which numerous cycles of conformational rearrangement are required to find a mutual orientation of substrates that is optimal for the chemical reaction.
Keywords: adenylate kinase; enzyme dynamics; single-molecule fluorescence.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
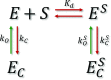
References
-
- Changeux JP. Allostery and the Monod-Wyman-Changeux model after 50 years. Annu Rev Biophys. 2012;41:103–133. - PubMed
-
- Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. Annu Rev Biochem. 2006;75:519–541. - PubMed
-
- Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

