IMPPAT: A curated database of Indian Medicinal Plants, Phytochemistry And Therapeutics
- PMID: 29531263
- PMCID: PMC5847565
- DOI: 10.1038/s41598-018-22631-z
IMPPAT: A curated database of Indian Medicinal Plants, Phytochemistry And Therapeutics
Abstract
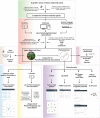
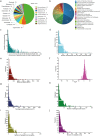
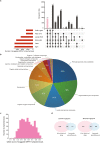
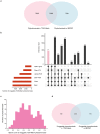
Phytochemicals of medicinal plants encompass a diverse chemical space for drug discovery. India is rich with a flora of indigenous medicinal plants that have been used for centuries in traditional Indian medicine to treat human maladies. A comprehensive online database on the phytochemistry of Indian medicinal plants will enable computational approaches towards natural product based drug discovery. In this direction, we present, IMPPAT, a manually curated database of 1742 Indian Medicinal Plants, 9596 Phytochemicals, And 1124 Therapeutic uses spanning 27074 plant-phytochemical associations and 11514 plant-therapeutic associations. Notably, the curation effort led to a non-redundant in silico library of 9596 phytochemicals with standard chemical identifiers and structure information. Using cheminformatic approaches, we have computed the physicochemical, ADMET (absorption, distribution, metabolism, excretion, toxicity) and drug-likeliness properties of the IMPPAT phytochemicals. We show that the stereochemical complexity and shape complexity of IMPPAT phytochemicals differ from libraries of commercial compounds or diversity-oriented synthesis compounds while being similar to other libraries of natural products. Within IMPPAT, we have filtered a subset of 960 potential druggable phytochemicals, of which majority have no significant similarity to existing FDA approved drugs, and thus, rendering them as good candidates for prospective drugs. IMPPAT database is openly accessible at: https://cb.imsc.res.in/imppat .
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

