Rapid sexual and genomic isolation in sympatric Drosophila without reproductive character displacement
- PMID: 29531700
- PMCID: PMC5838044
- DOI: 10.1002/ece3.3893
Rapid sexual and genomic isolation in sympatric Drosophila without reproductive character displacement
Abstract
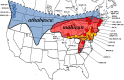
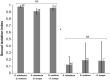
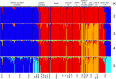
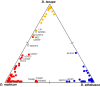
The rapid evolution of sexual isolation in sympatry has long been associated with reinforcement (i.e., selection to avoid maladaptive hybridization). However, there are many species pairs in sympatry that have evolved rapid sexual isolation without known costs to hybridization. A major unresolved question is what evolutionary processes are involved in driving rapid speciation in such cases. Here, we focus on one such system; the Drosophila athabasca species complex, which is composed of three partially sympatric and interfertile semispecies: WN, EA, and EB. To study speciation in this species complex, we assayed sexual and genomic isolation within and between these semispecies in both sympatric and allopatric populations. First, we found no evidence of reproductive character displacement (RCD) in sympatric zones compared to distant allopatry. Instead, semispecies were virtually completely sexually isolated from each other across their entire ranges. Moreover, using spatial approaches and coalescent demographic simulations, we detected either zero or only weak heterospecific gene flow in sympatry. In contrast, within each semispecies we found only random mating and little population genetic structure, except between highly geographically distant populations. Finally, we determined that speciation in this system is at least an order of magnitude older than previously assumed, with WN diverging first, around 200K years ago, and EA and EB diverging 100K years ago. In total, these results suggest that these semispecies should be given full species status and we adopt new nomenclature: WN-D. athabasca, EA-D. mahican, and EB-D. lenape. While the lack of RCD in sympatry and interfertility do not support reinforcement, we discuss what additional evidence is needed to further decipher the mechanisms that caused rapid speciation in this species complex.
Keywords: gene flow; genomic divergence; isolation by distance; population structure; range expansions; reproductive character displacement; secondary contact; sexual isolation.
Figures




References
-
- Barluenga, M. , Stölting, K. N. , Salzburger, W. , Muschick, M. , & Meyer, A. (2006). Sympatric speciation in Nicaraguan crater lake cichlid fish. Nature, 439(7077), 719 https://doi.org/10.1038/nature04325 - DOI - PubMed
-
- Beckenbach, A. T. , Wei, Y. W. , & Liu, H. (1993). Relationships in the Drosophila obscura species group, inferred from mitochondrial cytochrome oxidase II sequences. Molecular Biology and Evolution, 10, 619–634. - PubMed
-
- Brubaker, L. B. , Anderson, P. M. , Edwards, M. E. , & Lozhkin, A. V. (2005). Beringia as a glacial refugium for boreal trees and shrubs: New perspectives from mapped pollen data. Journal of Biogeography, 32(5), 833–848. https://doi.org/10.1111/j.1365-2699.2004.01203.x - DOI
-
- Bush, G. L. (1969). Sympatric host race formation and speciation in frugivorous flies of the genus Rhagoletis (Diptera, Tephritidae). Evolution, 23(2), 237–251. https://doi.org/10.1111/j.1558-5646.1969.tb03508.x - DOI - PubMed
-
- Butlin, R. K. (1995). Reinforcement—An idea evolving. Trends in Ecology & Evolution, 10(11), 432–434. https://doi.org/10.1016/S0169-5347(00)89173-9 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

