Chemical Space Expansion of Bromodomain Ligands Guided by in Silico Virtual Couplings (AutoCouple)
- PMID: 29532017
- PMCID: PMC5833004
- DOI: 10.1021/acscentsci.7b00401
Chemical Space Expansion of Bromodomain Ligands Guided by in Silico Virtual Couplings (AutoCouple)
Abstract
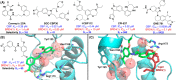
Expanding the chemical space and simultaneously ensuring synthetic accessibility is of upmost importance, not only for the discovery of effective binders for novel protein classes but, more importantly, for the development of compounds against hard-to-drug proteins. Here, we present AutoCouple, a de novo approach to computational ligand design focused on the diversity-oriented generation of chemical entities via virtual couplings. In a benchmark application, chemically diverse compounds with low-nanomolar potency for the CBP bromodomain and high selectivity against the BRD4(1) bromodomain were achieved by the synthesis of about 50 derivatives of the original fragment. The binding mode was confirmed by X-ray crystallography, target engagement in cells was demonstrated, and antiproliferative activity was showcased in three cancer cell lines. These results reveal AutoCouple as a useful in silico coupling method to expand the chemical space in hit optimization campaigns resulting in potent, selective, and cell permeable bromodomain ligands.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
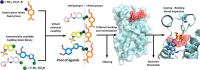

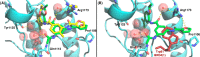

Similar articles
-
Design, synthesis, and biological evaluation of tetrahydroquinolin derivatives as potent inhibitors of CBP bromodomain.Bioorg Chem. 2020 Aug;101:103991. doi: 10.1016/j.bioorg.2020.103991. Epub 2020 Jun 2. Bioorg Chem. 2020. PMID: 32559581
-
Fragment-Based Design of Selective Nanomolar Ligands of the CREBBP Bromodomain.J Med Chem. 2016 Feb 25;59(4):1350-6. doi: 10.1021/acs.jmedchem.5b00172. Epub 2015 Jul 15. J Med Chem. 2016. PMID: 26043365
-
Virtual screen to NMR (VS2NMR): Discovery of fragment hits for the CBP bromodomain.Bioorg Med Chem Lett. 2017 Jun 1;27(11):2472-2478. doi: 10.1016/j.bmcl.2017.04.001. Epub 2017 Apr 4. Bioorg Med Chem Lett. 2017. PMID: 28410781
-
Fragment-based in silico screening of bromodomain ligands.Drug Discov Today Technol. 2016 Mar;19:81-90. doi: 10.1016/j.ddtec.2016.06.003. Epub 2016 Jul 21. Drug Discov Today Technol. 2016. PMID: 27769362 Review.
-
Structure-guided fragment screening for lead discovery.Curr Opin Drug Discov Devel. 2004 Jul;7(4):404-10. Curr Opin Drug Discov Devel. 2004. PMID: 15338949 Review.
Cited by
-
Selective CBP/EP300 Bromodomain Inhibitors: Novel Epigenetic Tools to Counter TNF-α-Driven Inflammation.JACS Au. 2025 Jun 5;5(6):2491-2499. doi: 10.1021/jacsau.5c00085. eCollection 2025 Jun 23. JACS Au. 2025. PMID: 40575322 Free PMC article.
-
ChemoDOTS: a web server to design chemistry-driven focused libraries.Nucleic Acids Res. 2024 Jul 5;52(W1):W461-W468. doi: 10.1093/nar/gkae326. Nucleic Acids Res. 2024. PMID: 38686808 Free PMC article.
-
Binding mode information improves fragment docking.J Cheminform. 2019 Mar 22;11(1):24. doi: 10.1186/s13321-019-0346-7. J Cheminform. 2019. PMID: 30903304 Free PMC article.
-
ASD2023: towards the integrating landscapes of allosteric knowledgebase.Nucleic Acids Res. 2024 Jan 5;52(D1):D376-D383. doi: 10.1093/nar/gkad915. Nucleic Acids Res. 2024. PMID: 37870448 Free PMC article.
-
CReM: chemically reasonable mutations framework for structure generation.J Cheminform. 2020 Apr 22;12(1):28. doi: 10.1186/s13321-020-00431-w. J Cheminform. 2020. PMID: 33430959 Free PMC article.
References
-
- Fink T.; Reymond J.-L. Virtual Exploration of the Chemical Universe up to 11 Atoms of C, N, O, F: Assembly of 26.4 Million Structures (110.9 Million Stereoisomers) and Analysis for New Ring Systems, Stereochemistry, Physicochemical Properties, Compound Classes, and Drug Discovery. J. Chem. Inf. Model. 2007, 47, 342–353. 10.1021/ci600423u. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

