Identification of differentially expressed genes and biological pathways in bladder cancer
- PMID: 29532898
- PMCID: PMC5928619
- DOI: 10.3892/mmr.2018.8711
Identification of differentially expressed genes and biological pathways in bladder cancer
Abstract
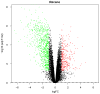
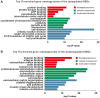
The purpose of the present study was to identify key genes and investigate the related molecular mechanisms of bladder cancer (BC) progression. From the Gene Expression Omnibus database, the gene expression dataset GSE7476 was downloaded, which contained 43 BC samples and 12 normal bladder tissues. GSE7476 was analyzed to screen the differentially expressed genes (DEGs). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed for the DEGs using the DAVID database, and a protein‑protein interaction (PPI) network was then constructed using Cytoscape software. The results of the GO analysis showed that the upregulated DEGs were significantly enriched in cell division, nucleoplasm and protein binding, while the downregulated DEGs were significantly enriched in 'extracellular matrix organization', 'proteinaceous extracellular matrix' and 'heparin binding'. The results of the KEGG pathway analysis showed that the upregulated DEGs were significantly enriched in the 'cell cycle', whereas the downregulated DEGs were significantly enriched in 'complement and coagulation cascades'. JUN, cyclin‑dependent kinase 1, FOS, PCNA, TOP2A, CCND1 and CDH1 were found to be hub genes in the PPI network. Sub‑networks revealed that these gene were enriched in significant pathways, including the 'cell cycle' signaling pathway and 'PI3K‑Akt signaling pathway'. In summary, the present study identified DEGs and key target genes in the progression of BC, providing potential molecular targets and diagnostic biomarkers for the treatment of BC.
Keywords: bioinformatics analysis; bladder cancer; differentially expressed genes; enrichment analysis; protein-protein interaction.
Figures


Similar articles
-
Identification of candidate biomarkers and pathways associated with SCLC by bioinformatics analysis.Mol Med Rep. 2018 Aug;18(2):1538-1550. doi: 10.3892/mmr.2018.9095. Epub 2018 May 29. Mol Med Rep. 2018. PMID: 29845250 Free PMC article.
-
Screening key genes and signaling pathways in colorectal cancer by integrated bioinformatics analysis.Mol Med Rep. 2019 Aug;20(2):1259-1269. doi: 10.3892/mmr.2019.10336. Epub 2019 Jun 4. Mol Med Rep. 2019. PMID: 31173250 Free PMC article.
-
Bioinformatics analyses of significant genes, related pathways and candidate prognostic biomarkers in glioblastoma.Mol Med Rep. 2018 Nov;18(5):4185-4196. doi: 10.3892/mmr.2018.9411. Epub 2018 Aug 21. Mol Med Rep. 2018. PMID: 30132538 Free PMC article.
-
Integrated bioinformatics analysis reveals key candidate genes and pathways in breast cancer.Mol Med Rep. 2018 Jun;17(6):8091-8100. doi: 10.3892/mmr.2018.8895. Epub 2018 Apr 19. Mol Med Rep. 2018. PMID: 29693125 Free PMC article.
-
Wnt signalling pathway in bladder cancer.Cell Signal. 2021 Mar;79:109886. doi: 10.1016/j.cellsig.2020.109886. Epub 2020 Dec 17. Cell Signal. 2021. PMID: 33340660 Review.
Cited by
-
A two-way rectification method for identifying differentially expressed genes by maximizing the co-function relationship.BMC Genomics. 2021 Jun 25;22(Suppl 1):471. doi: 10.1186/s12864-021-07772-2. BMC Genomics. 2021. PMID: 34171992 Free PMC article.
-
Upregulation of ASPM, BUB1B and SPDL1 in tumor tissues predicts poor survival in patients with pancreatic ductal adenocarcinoma.Oncol Lett. 2020 Apr;19(4):3307-3315. doi: 10.3892/ol.2020.11414. Epub 2020 Feb 19. Oncol Lett. 2020. PMID: 32218868 Free PMC article.
-
Gene network screening of bladder cancer via modular analysis.Transl Cancer Res. 2021 Feb;10(2):1043-1052. doi: 10.21037/tcr-20-2822. Transl Cancer Res. 2021. PMID: 35116431 Free PMC article.
-
Exploiting protein family and protein network data to identify novel drug targets for bladder cancer.Oncotarget. 2022 Jan 12;13:105-117. doi: 10.18632/oncotarget.28175. eCollection 2022. Oncotarget. 2022. PMID: 35035776 Free PMC article.
-
Down-expression of GOLM1 enhances the chemo-sensitivity of cervical cancer to methotrexate through modulation of the MMP13/EMT axis.Am J Cancer Res. 2018 Jun 1;8(6):964-980. eCollection 2018. Am J Cancer Res. 2018. PMID: 30034935 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

