NuRD and CAF-1-mediated silencing of the D4Z4 array is modulated by DUX4-induced MBD3L proteins
- PMID: 29533181
- PMCID: PMC5849414
- DOI: 10.7554/eLife.31023
NuRD and CAF-1-mediated silencing of the D4Z4 array is modulated by DUX4-induced MBD3L proteins
Abstract
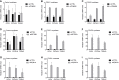
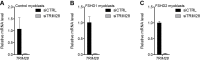
The DUX4 transcription factor is encoded by a retrogene embedded in each unit of the D4Z4 macrosatellite repeat. DUX4 is normally expressed in the cleavage-stage embryo, whereas chromatin repression prevents DUX4 expression in most somatic tissues. Failure of this repression causes facioscapulohumeral muscular dystrophy (FSHD) due to mis-expression of DUX4 in skeletal muscle. In this study, we used CRISPR/Cas9 engineered chromatin immunoprecipitation (enChIP) locus-specific proteomics to characterize D4Z4-associated proteins. These and other approaches identified the Nucleosome Remodeling Deacetylase (NuRD) and Chromatin Assembly Factor 1 (CAF-1) complexes as necessary for DUX4 repression in human skeletal muscle cells and induced pluripotent stem (iPS) cells. Furthermore, DUX4-induced expression of MBD3L proteins partly relieved this repression in FSHD muscle cells. Together, these findings identify NuRD and CAF-1 as mediators of DUX4 chromatin repression and suggest a mechanism for the amplification of DUX4 expression in FSHD muscle cells.
Keywords: CAF-1; D4Z4; DUX4; MBD3L; NuRD; chromosomes; facioscapulohumeral dystrophy; genes; human.
© 2018, Campbell et al.
Conflict of interest statement
AC, SS, SJ, JL, RR, RT, Sv, ST No competing interests declared
Figures



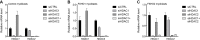
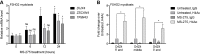
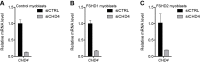
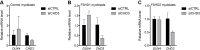
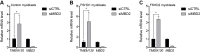

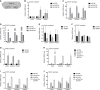
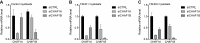
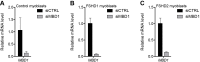

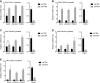
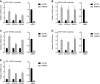
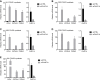
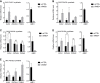
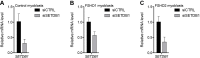
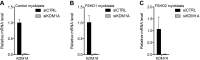


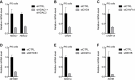
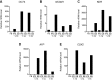
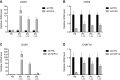
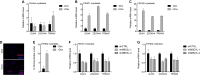

References
-
- Balog J, Thijssen PE, Shadle S, Straasheijm KR, van der Vliet PJ, Krom YD, van den Boogaard ML, de Jong A, F Lemmers RJ, Tawil R, Tapscott SJ, van der Maarel SM. Increased DUX4 expression during muscle differentiation correlates with decreased SMCHD1 protein levels at D4Z4. Epigenetics. 2015;10:1133–1142. doi: 10.1080/15592294.2015.1113798. - DOI - PMC - PubMed
-
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermüller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, Drenkow J, Bell I, Zhao X, Srinivasan KG, Sung WK, Ooi HS, Chiu KP, Foissac S, Alioto T, Brent M, Pachter L, Tress ML, Valencia A, Choo SW, Choo CY, Ucla C, Manzano C, Wyss C, Cheung E, Clark TG, Brown JB, Ganesh M, Patel S, Tammana H, Chrast J, Henrichsen CN, Kai C, Kawai J, Nagalakshmi U, Wu J, Lian Z, Lian J, Newburger P, Zhang X, Bickel P, Mattick JS, Carninci P, Hayashizaki Y, Weissman S, Hubbard T, Myers RM, Rogers J, Stadler PF, Lowe TM, Wei CL, Ruan Y, Struhl K, Gerstein M, Antonarakis SE, Fu Y, Green ED, Karaöz U, Siepel A, Taylor J, Liefer LA, Wetterstrand KA, Good PJ, Feingold EA, Guyer MS, Cooper GM, Asimenos G, Dewey CN, Hou M, Nikolaev S, Montoya-Burgos JI, Löytynoja A, Whelan S, Pardi F, Massingham T, Huang H, Zhang NR, Holmes I, Mullikin JC, Ureta-Vidal A, Paten B, Seringhaus M, Church D, Rosenbloom K, Kent WJ, Stone EA, Batzoglou S, Goldman N, Hardison RC, Haussler D, Miller W, Sidow A, Trinklein ND, Zhang ZD, Barrera L, Stuart R, King DC, Ameur A, Enroth S, Bieda MC, Kim J, Bhinge AA, Jiang N, Liu J, Yao F, Vega VB, Lee CW, Ng P, Shahab A, Yang A, Moqtaderi Z, Zhu Z, Xu X, Squazzo S, Oberley MJ, Inman D, Singer MA, Richmond TA, Munn KJ, Rada-Iglesias A, Wallerman O, Komorowski J, Fowler JC, Couttet P, Bruce AW, Dovey OM, Ellis PD, Langford CF, Nix DA, Euskirchen G, Hartman S, Urban AE, Kraus P, Van Calcar S, Heintzman N, Kim TH, Wang K, Qu C, Hon G, Luna R, Glass CK, Rosenfeld MG, Aldred SF, Cooper SJ, Halees A, Lin JM, Shulha HP, Zhang X, Xu M, Haidar JN, Yu Y, Ruan Y, Iyer VR, Green RD, Wadelius C, Farnham PJ, Ren B, Harte RA, Hinrichs AS, Trumbower H, Clawson H, Hillman-Jackson J, Zweig AS, Smith K, Thakkapallayil A, Barber G, Kuhn RM, Karolchik D, Armengol L, Bird CP, de Bakker PI, Kern AD, Lopez-Bigas N, Martin JD, Stranger BE, Woodroffe A, Davydov E, Dimas A, Eyras E, Hallgrímsdóttir IB, Huppert J, Zody MC, Abecasis GR, Estivill X, Bouffard GG, Guan X, Hansen NF, Idol JR, Maduro VV, Maskeri B, McDowell JC, Park M, Thomas PJ, Young AC, Blakesley RW, Muzny DM, Sodergren E, Wheeler DA, Worley KC, Jiang H, Weinstock GM, Gibbs RA, Graves T, Fulton R, Mardis ER, Wilson RK, Clamp M, Cuff J, Gnerre S, Jaffe DB, Chang JL, Lindblad-Toh K, Lander ES, Koriabine M, Nefedov M, Osoegawa K, Yoshinaga Y, Zhu B, de Jong PJ, ENCODE Project Consortium. NISC Comparative Sequencing Program. Baylor College of Medicine Human Genome Sequencing Center. Washington University Genome Sequencing Center. Broad Institute. Children's Hospital Oakland Research Institute Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 NS069539/NS/NINDS NIH HHS/United States
- T32GM007270/GM/NIGMS NIH HHS/United States
- T32 GM007270/GM/NIGMS NIH HHS/United States
- P01NS069539/NS/NINDS NIH HHS/United States
- R01 AR045203/AR/NIAMS NIH HHS/United States
- T32HG000035/HG/NHGRI NIH HHS/United States
- T32 CA009657/CA/NCI NIH HHS/United States
- T32 GM007266/GM/NIGMS NIH HHS/United States
- T32HG00035/HG/NHGRI NIH HHS/United States
- R01AR045203/AR/NIAMS NIH HHS/United States
- T32CA009657/CA/NCI NIH HHS/United States
- T32 HD007183/HD/NICHD NIH HHS/United States
- T32 HG000035/HG/NHGRI NIH HHS/United States
- R01AR066248/AR/NIAMS NIH HHS/United States
- R01 AR066248/AR/NIAMS NIH HHS/United States
- T32HD007183/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

